SecStrAnnotator:UserManual: Difference between revisions
No edit summary |
No edit summary |
||
| (10 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
[[File:SecStrAnnotator-annotated-2nnj.png | thumb | right | 400px | Structure of human CYP 2C8 (PDB ID 2nnj) with annotation of the major helices.]] | |||
'''SecStrAnnotator''' is a command line tool for annotation (naming) of '''secondary structure elements (SSEs)''', namely helices and β-strands, in protein structures. It is template-based, meaning that a structurally similar template protein with annotated SSEs is a part of the input. | '''SecStrAnnotator''' is a command line tool for annotation (naming) of '''secondary structure elements (SSEs)''', namely helices and β-strands, in protein structures. It is template-based, meaning that a structurally similar template protein with annotated SSEs is a part of the input. | ||
'''SecStrAnnotator''' alone can be used for '''one-to-one annotation'''. This means that it takes an annotated template protein ''T'' and a query protein ''Q'', it finds SSEs in the query protein ''Q'', and annotates the SSEs | '''SecStrAnnotator''' alone can be used for '''one-to-one annotation'''. This means that it takes an annotated template protein ''T'' and a query protein ''Q'', it finds SSEs in the query protein ''Q'', and annotates the SSEs in a way that best reflects the annotation of the template ''T''. | ||
| Line 9: | Line 11: | ||
'''SecStrAPI''' provides precomputed SSE annotations (currently only for Cytochrome P450 family). | |||
'''[http://sestra.ncbr.muni.cz SecStrAnnotator Online]''' is a web application allowing you to annotate SSEs without installing the full version of SecStrAnnotator. | |||
'''NEWS:''' | '''NEWS:''' | ||
'''SecStrAnnotator 2 | '''SecStrAnnotator 2''' is now available. There are a few basic differences from SecStrAnnotator 1.0: | ||
* SecStrAnnotator 2 | * SecStrAnnotator 2 is based on .NET Core and it is run using .NET Core Runtime (<code>dotnet</code>). This provides the same behaviour in each operating system. | ||
* The supported structural file format is now mmCIF (PDB file format is not supported anymore). Chain and residue numbering is based on label_* columns in the mmCIF files, not auth_* columns! | * The supported structural file format is now mmCIF (PDB file format is not supported anymore). Chain and residue numbering is based on label_* columns in the mmCIF files, not auth_* columns! | ||
| Line 29: | Line 34: | ||
===[[SecStrAnnotator:OneToMany | Annotation of a whole protein family (one-to-many)]]=== | ===[[SecStrAnnotator:OneToMany | Annotation of a whole protein family (one-to-many)]]=== | ||
---- | ---- | ||
===[[SecStrAnnotator:Analysis | Analysis of family annotation]]=== | ===[[SecStrAnnotator:Analysis | Analysis of family annotation (case study: Cytochromes P450)]]=== | ||
---- | |||
===[[SecStrAnnotator:SecStrAPI | SecStrAPI]]=== | |||
---- | ---- | ||
===[[SecStrAnnotator:Download | Download]]=== | ===[[SecStrAnnotator:Download | Download]]=== | ||
Latest revision as of 21:44, 22 March 2021
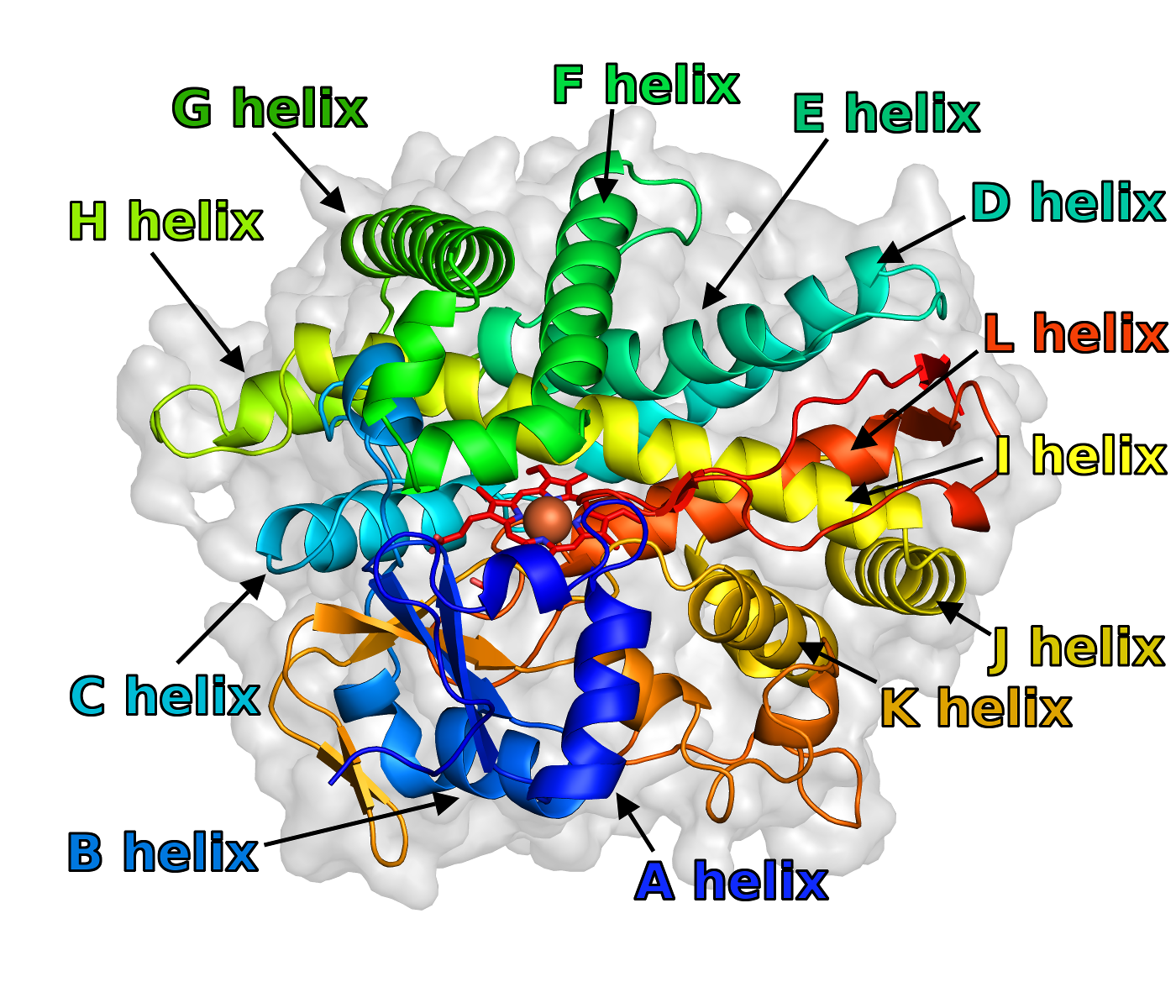
SecStrAnnotator is a command line tool for annotation (naming) of secondary structure elements (SSEs), namely helices and β-strands, in protein structures. It is template-based, meaning that a structurally similar template protein with annotated SSEs is a part of the input.
SecStrAnnotator alone can be used for one-to-one annotation. This means that it takes an annotated template protein T and a query protein Q, it finds SSEs in the query protein Q, and annotates the SSEs in a way that best reflects the annotation of the template T.
SecStrAnnotator Suite is a set of additional scripts which can be used for one-to-many annotation. This means that we have a whole protein family, and one of its members serves as the template T. The result is the annotation of all other members of the family. The suite also provides Python and R scripts for a further analysis of the annotation results.
SecStrAPI provides precomputed SSE annotations (currently only for Cytochrome P450 family).
SecStrAnnotator Online is a web application allowing you to annotate SSEs without installing the full version of SecStrAnnotator.
NEWS:
SecStrAnnotator 2 is now available. There are a few basic differences from SecStrAnnotator 1.0:
- SecStrAnnotator 2 is based on .NET Core and it is run using .NET Core Runtime (
dotnet). This provides the same behaviour in each operating system.
- The supported structural file format is now mmCIF (PDB file format is not supported anymore). Chain and residue numbering is based on label_* columns in the mmCIF files, not auth_* columns!
If you found SecStrAnnotator helpful, please cite:
Midlik A, Hutařová Vařeková I, Hutař J, Moturu TR, Navrátilová V, Koča J, Berka K, Svobodová Vařeková R (2019) Automated family-wide annotation of secondary structure elements. In: Kister AE (ed) Protein supersecondary structures: Methods and protocols, Humana Press. ISBN 978-1-4939-9160-0.

|
SecStrAnnotator Suite is a part of services provided by ELIXIR – European research infrastructure for biological information. For other services provided by ELIXIR's Czech Republic Node visit www.elixir-czech.cz/services. |