SecStrAnnotator:SecStrAPI
Appearance
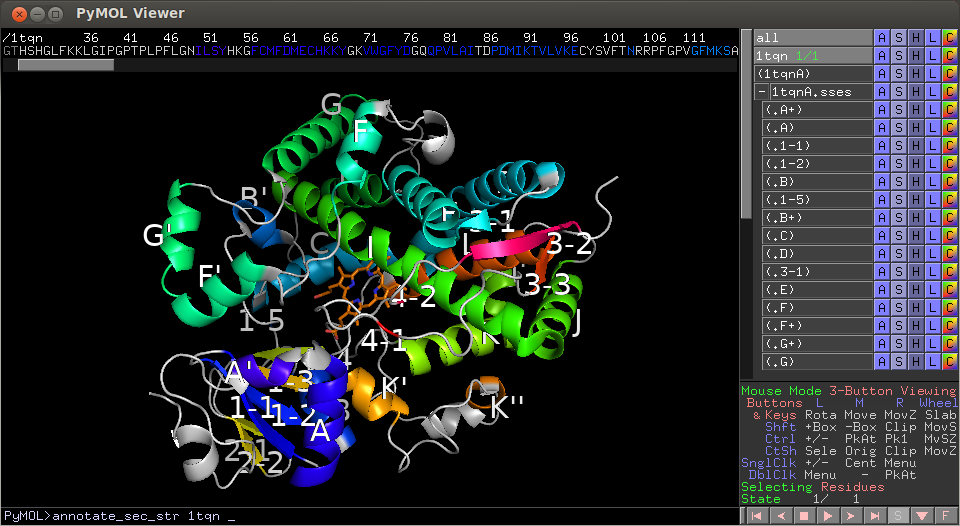
> run https://webchem.ncbr.muni.cz/API/SecStr/Downloads/secstrapi_plugin.py > annotate_sec_str 1tqnThe SecStrAPI allows programmatic access to precomputed secondary structure annotations. The annotations from SecStrAPI can also be easily visualized in PyMOL using SecStrAPI Plugin.
Requests
[edit]-
The annotation for a particular PDB entry is accessible by:
https://webchem.ncbr.muni.cz/API/SecStr/Annotation/{pdbid}
Example:https://webchem.ncbr.muni.cz/API/SecStr/Annotation/1tqn -
The list of available PDB entries and the annotated domains in each entry is accessible by:
https://webchem.ncbr.muni.cz/API/SecStr/List/Annotation
SecStrAPI format
[edit]The annotations are provided in SecStrAPI JSON format, demonstrated and explained below.
{
"api_version": "1.0",
"annotations": {
"1tqn": { # PDB ID
"1tqnA": { # domain ID (unique domain identifier in SecStrAPI)
"pdb": "1tqn", # PDB ID
"chain_id": "A", # mmcif-style chain identifier (label_asym_id)
"ranges": ":", # mmcif-style residue range (label_seq_id), : stands for the whole chain
"auth_chain_id": "A", # pdb-style chain identifier (auth_asym_id)
"auth_ranges": ":", # pdb-style residue range (auth_seq_id), : stands for the whole chain
"uniprot_id": "P08684", # UniProt accession number
"uniprot_name": "CP3A4_HUMAN", # UniProt mnemonic identifier
"domain_mappings": [ # list of mappings to source databases
{
"domain": "1tqnA00", # domain ID in the source database, if available
"source": "CATH", # name of source database
"family": "1.10.630.10", # family ID in the source database
"pdb": "1tqn", # PDB ID
"chain_id": "A", # mmcif-style chain identifier (label_asym_id) of the domain in the source database
"ranges": "7:478" # mmcif-style residue range (label_seq_id) of the domain in the source database (including the number before and after the colon)
},
{
"domain": null,
"source": "Pfam",
"family": "PF00067",
"pdb": "1tqn",
"chain_id": "A",
"ranges": "17:472"
}
],
"secondary_structure_elements": [ # list of annotated secondary structure elements (SSEs)
{
"label": "1-1", # SSE label
"chain_id": "A", # mmcif-style chain identifier (label_asym_id)
"start": 50, # mmcif-style residue number of the first residue (label_seq_id)
"end": 55, # mmcif-style residue number of the last residue (label_seq_id)
"auth_chain_id": "A", # pdb-style chain identifier (auth_asym_id)
"auth_start": 71, # pdb-style residue number of the first residue (auth_seq_id)
"auth_start_ins_code": "", # residue insertion code of the first residue (pdbx_PDB_ins_code)
"auth_end": 76, # pdb-style residue number of the last residue (auth_seq_id)
"auth_end_ins_code": "", # residue insertion code of the last residue (pdbx_PDB_ins_code)
"type": "e", # SSE type (h: helix, e: beta-strand)
"sheet_id": 1, # beta-sheet identifier (all strands in the same sheet have the same sheet_id)
"color": "s164", # pymol-style color assigned to this SSE (consistent through the whole family)
"metric_value": 2.5, # measure of deviation from the template
"confidence": "high", # confidence of the annotation (high: metric_value <= 10, medium: 10 < metric_value <= 20, low: metric_value > 20)
"sequence": "VWGFYD" # amino acid sequence of the SSE
},
{
"label": "1-2",
"chain_id": "A",
"start": 58,
"end": 63,
"auth_chain_id": "A",
"auth_start": 79,
"auth_start_ins_code": "",
"auth_end": 84,
"auth_end_ins_code": "",
"type": "e",
"sheet_id": 1,
"color": "s186",
"metric_value": 2.03,
"confidence": "high",
"sequence": "QPVLAI"
},
{
"label": "B",
"chain_id": "A",
"start": 66,
"end": 76,
"auth_chain_id": "A",
"auth_start": 87,
"auth_start_ins_code": "",
"auth_end": 97,
"auth_end_ins_code": "",
"type": "h",
"color": "s208",
"metric_value": 3.83,
"confidence": "high",
"sequence": "PDMIKTVLVKE"
}
],
"beta_connectivity": [ # list of connections between strands in beta-sheets
[
"1-1", # label of the first strand
"1-2", # label of the second strand
-1 # relative orientation of the strands (1: parallel, -1: antiparallel)
]
],
"rotation_matrix": [ # PyMOL object matrix used to align the domain to the template
0.0841438366722304,
-0.9907657680912565,
-0.10631560341088532,
-0.46321294579603234,
0.04653041207831938,
-0.10267083975128484,
0.99362649895048,
-0.12159261906614163,
-0.995366633709358,
-0.08855445467795255,
0.03746162109134595,
0.05273612685656731,
18.78404164503984,
23.992462979534775,
13.616655034021722,
1.0
],
"comment": "Automatic annotation for 1tqn based on 2NNJ template. Program was called with these parameters: /home/adam/Workspace/C#/SecStrAnnot2_data/SecStrAPI/CytochromesP450-20200128/structures 2NNJ,A,: 1tqn,A,: --soft --label2auth --maxmetric 25,0.5,0.5 --verbose Total value of used metric: 186.50"
}
}
}
}