SecStrAnnotator:Analysis: Difference between revisions
Created page with "Under construction <br style="clear:both" /> ---- Back to the main page" |
No edit summary |
||
| (24 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
SecStrAnnotator Suite provides scripts (Python, R) for batch annotation of the whole family and analysis of the annotation results. | |||
==Procedure== | |||
===Data preparation=== | |||
The directory <code>scripts/secstrapi_data_preparation/</code> contains a pipeline for annotating the whole protein family, including: | |||
* downloading the list of family members defined by CATH and Pfam, | |||
* downloading their structures, | |||
* selecting a non-redundant set, | |||
* annotation, | |||
* multiple sequence alignment for individual SSEs, | |||
* formatting into [[SecStrAnnotator:SecStrAPI#SecStrAPI_format | SecStrAPI format]], | |||
* formatting into TSV format for further analyses. | |||
The whole pipeline can be executed by <code>scripts/SecStrAPI_pipeline.py</code> | |||
Example usage: | |||
python3 scripts/SecStrAPI_pipeline.py scripts/SecStrAPI_pipeline_settings.json --resume | |||
Before running, modify the settings in <code>SecStrAPI_pipeline_settings.json</code> to set your family of interest, annotation template, data directory etc (see <code>README.txt</code> for more details). | |||
===Data analysis=== | |||
The directory <code>scripts/R_sec_str_anatomy_analysis/</code> contains a pipeline for statistical analysis of the annotation results on the whole protein family, including: | |||
* reading the annotation results from TSV, | |||
* generating plots, | |||
* performing statistical test to compare eukaryotic and bacterial structures (or any two sets of structures). | |||
Example usage: | |||
* Launch <code>rstudio</code> from the said directory | |||
* In <code>sec_str_anatomy.R</code>, set DATADIR to the path to your annotation data created in [[#Data preparation]] | |||
* In <code>sec_str_anatomy_settings.R</code>, modify the family-specific settings (list of helices and strands) | |||
* Run <code>sec_str_anatomy.R</code> line by line | |||
==Example case study: Cytochromes P450== | |||
===Data=== | |||
For the Cytochrome P450 family, structures of 1855 protein domains are available, located in 1012 PDB entries (updated on 7 July 2020). The analysis was performed on a non-redundant subset containing 183 protein domains. | |||
The data are available [https://doi.org/10.5281/zenodo.3939133 here] (structural files not included because of their size). | |||
===Occurrence of SSEs=== | |||
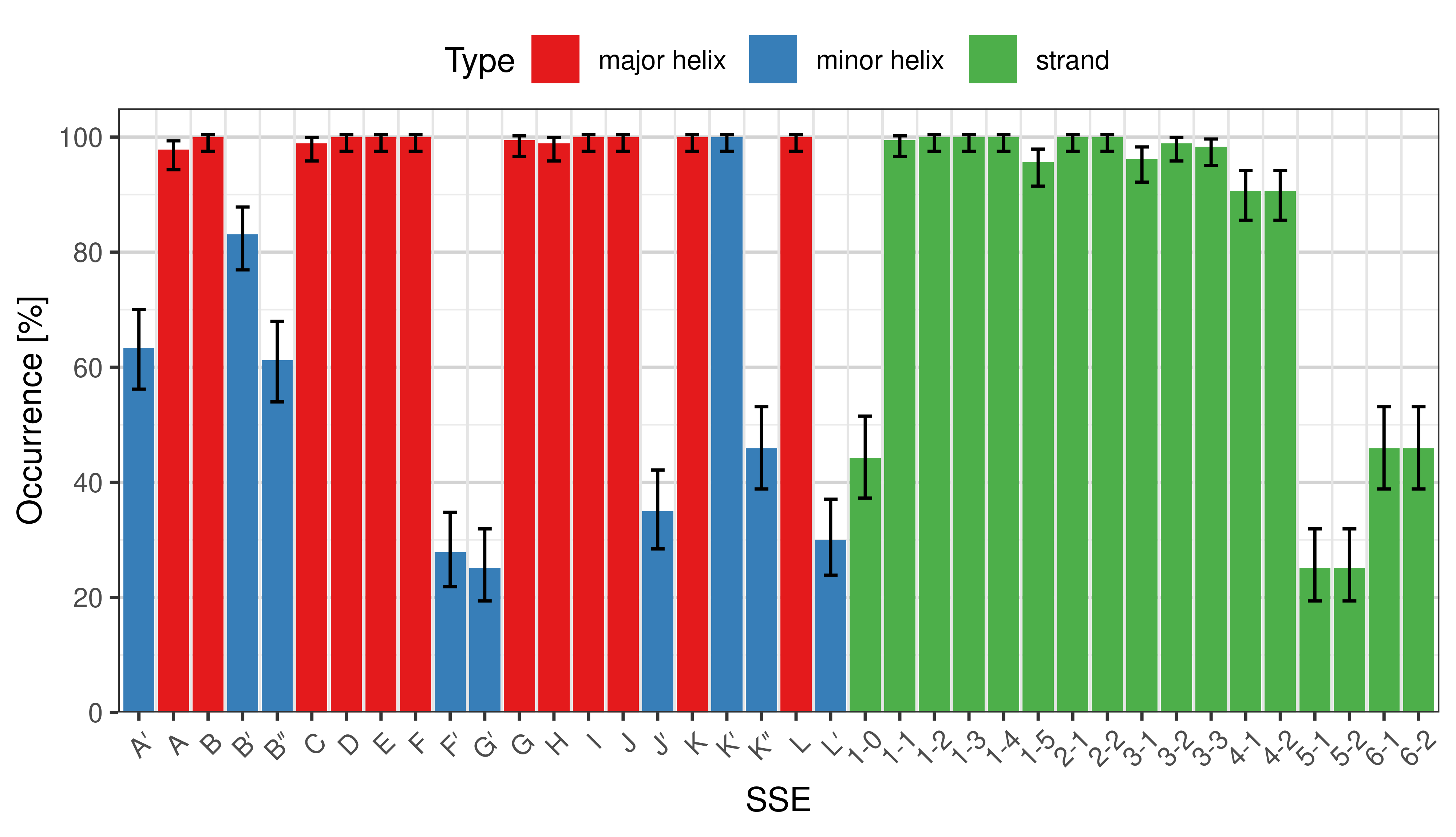
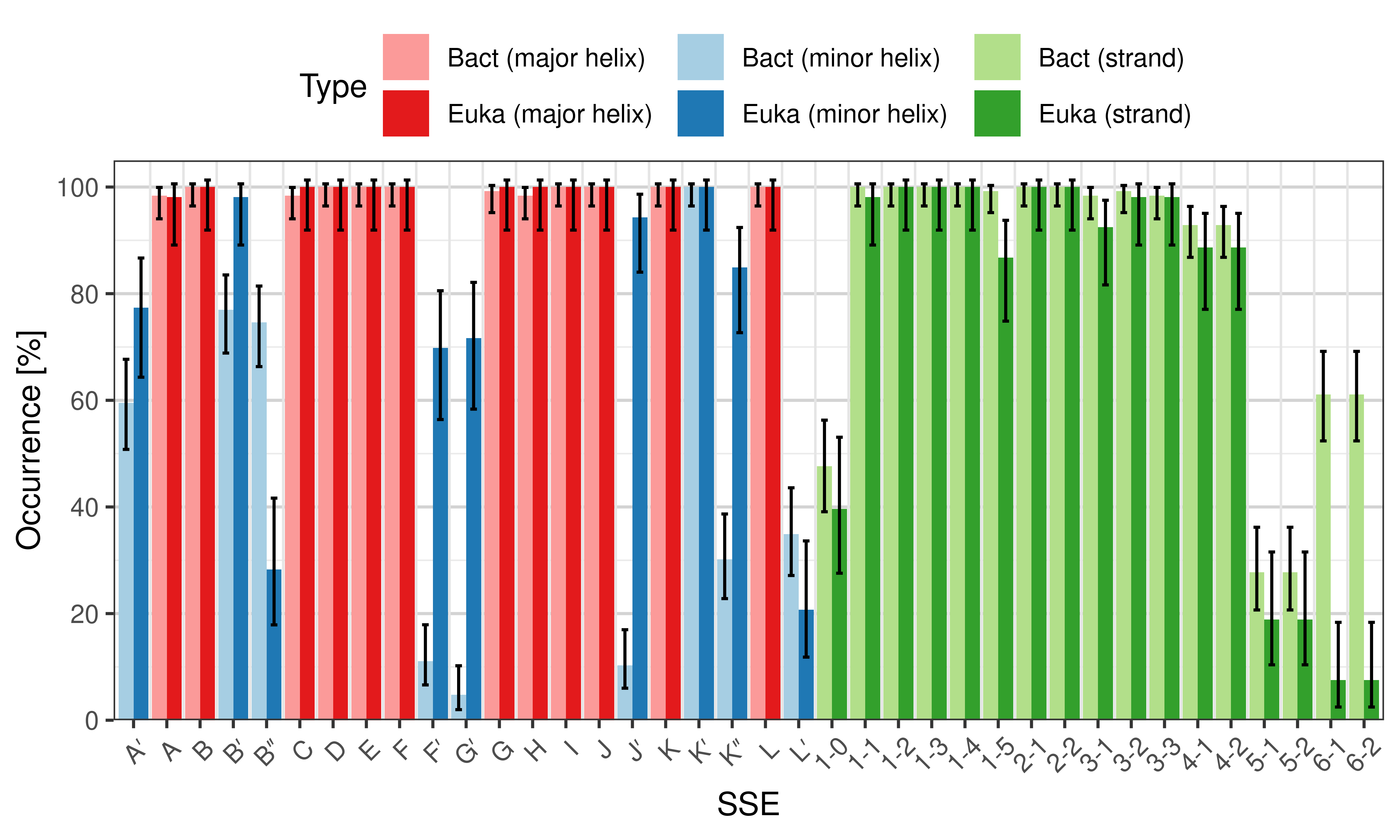
The ''occurrence'' describes in what percentage of the structures a particular SSE is present. | |||
<div><ul> | |||
<li style="display: inline-block;"> [[File:SecStrAnnotator-cyp-sse-occurrence.png | thumb | 500px | Occurrence of particular SSEs in the whole set.]] </li> | |||
<li style="display: inline-block;"> [[File:SecStrAnnotator-cyp-sse-occurrence-Bact-Euka.png | thumb | 500px | Occurrence of particular SSEs – comparison of bacterial and eukaryotic structures.]] </li> | |||
</ul></div> | |||
===Length of SSEs=== | |||
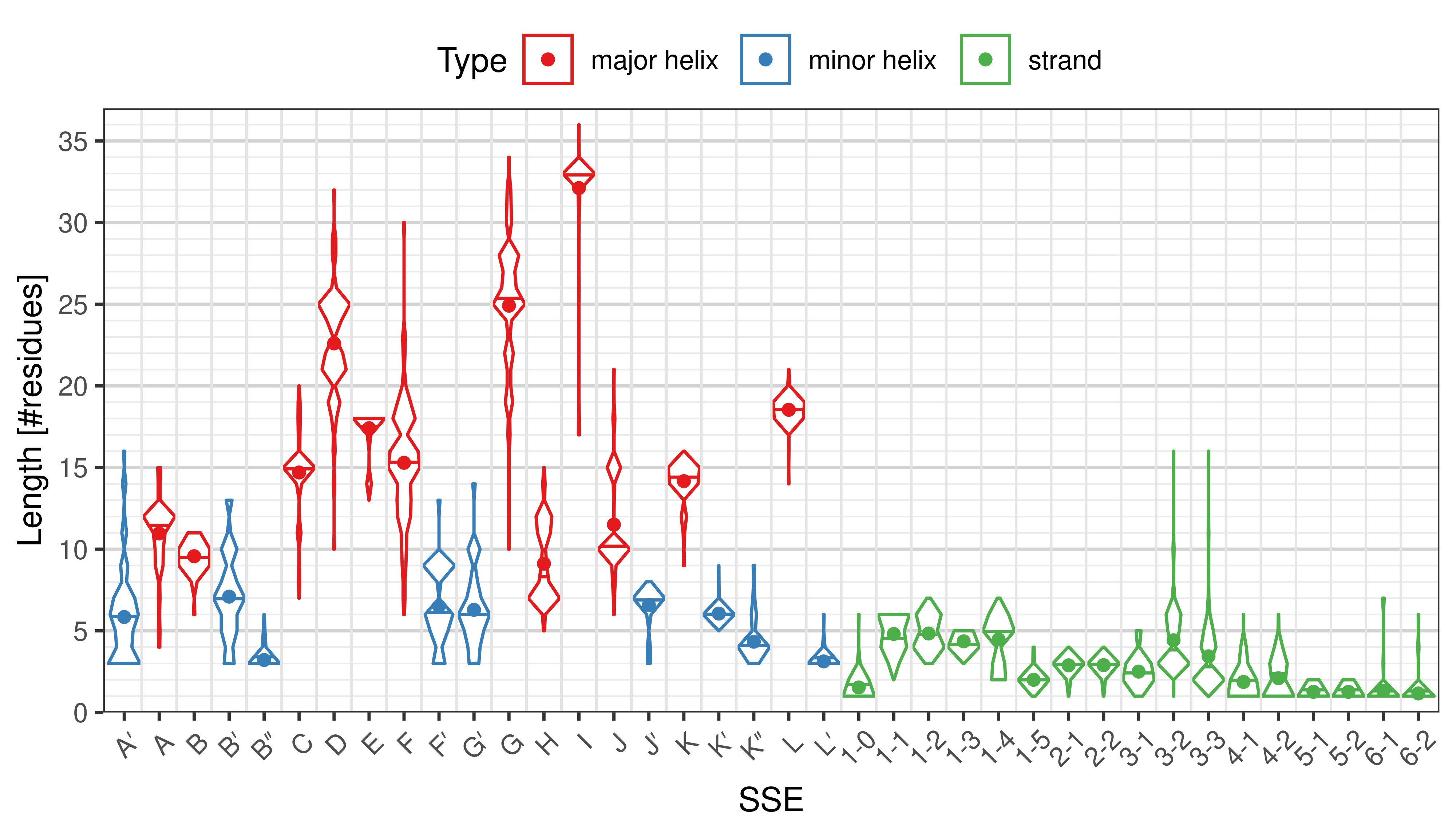
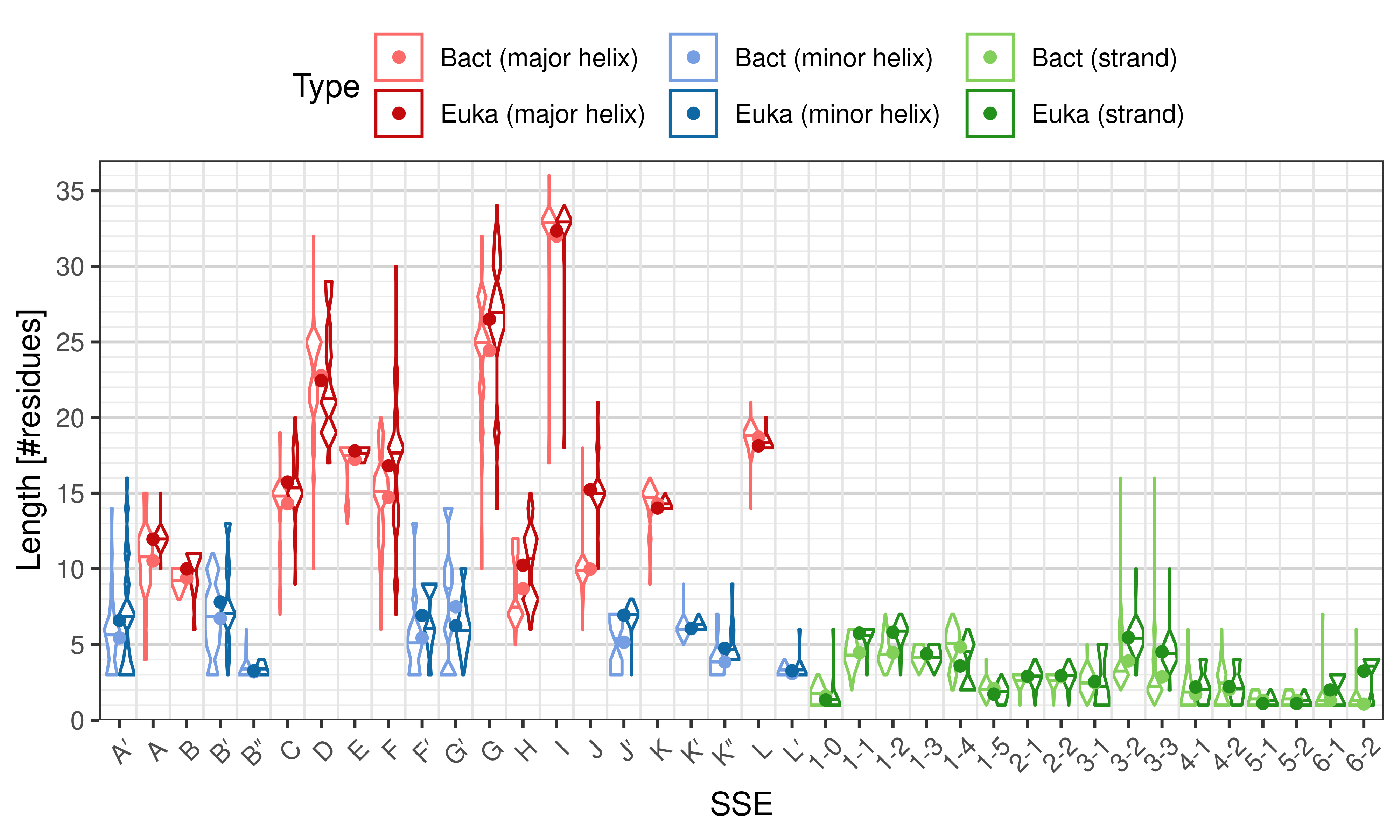
The ''length'' of an SSE is measured as the number of residues. The following violin plots show the distribution of length for each SSE. | |||
<div><ul> | |||
<li style="display: inline-block;"> [[File:SecStrAnnotator-cyp-sse-length.png | thumb | 500px | Length distribution of particular SSEs in the whole set. ]] </li> | |||
<li style="display: inline-block;"> [[File:SecStrAnnotator-cyp-sse-length-Bact-Euka.png | thumb | 500px | Length distribution of particular SSEs – comparison of bacterial and eukaryotic structures. ]] </li> | |||
</ul></div> | |||
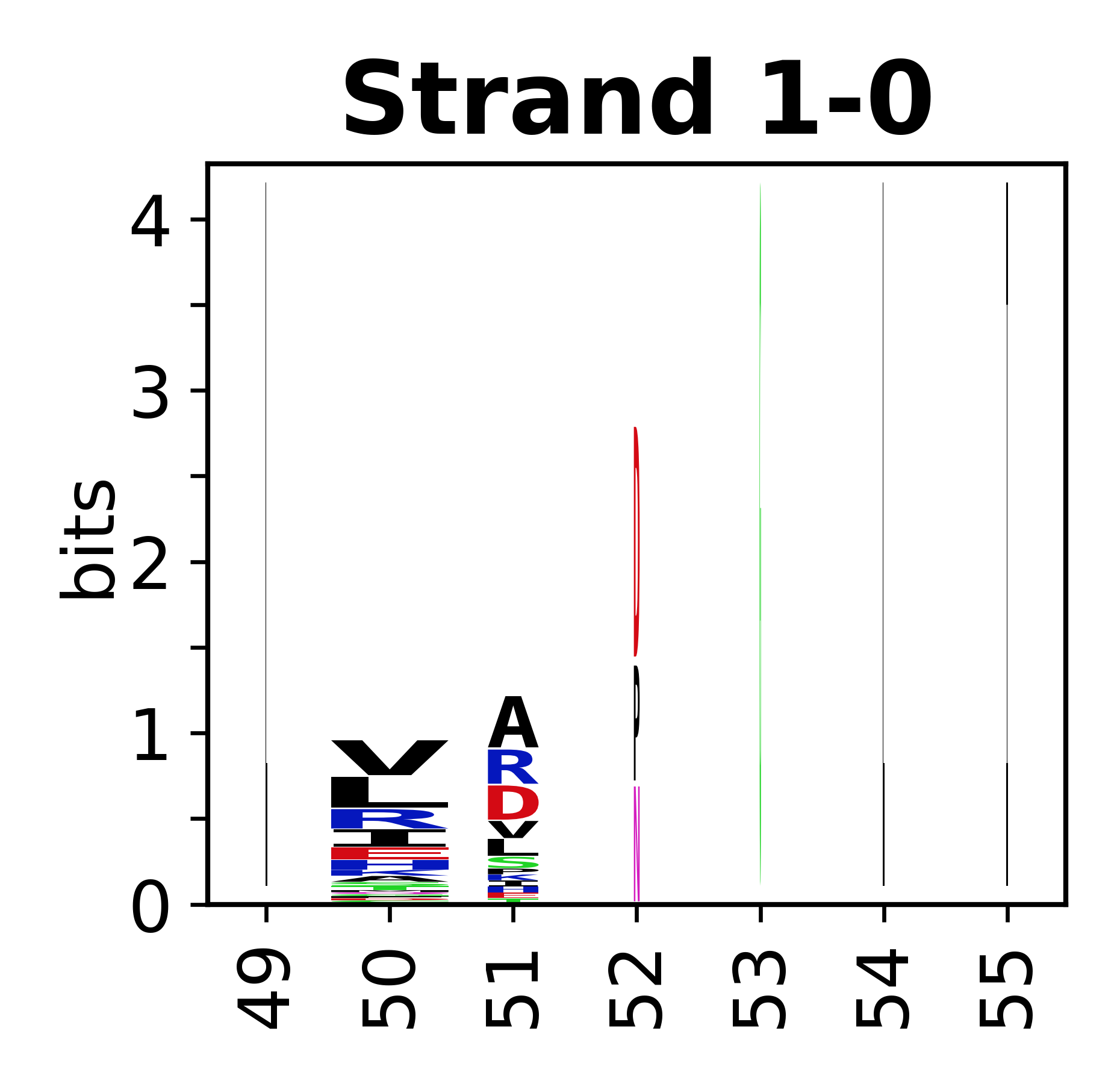
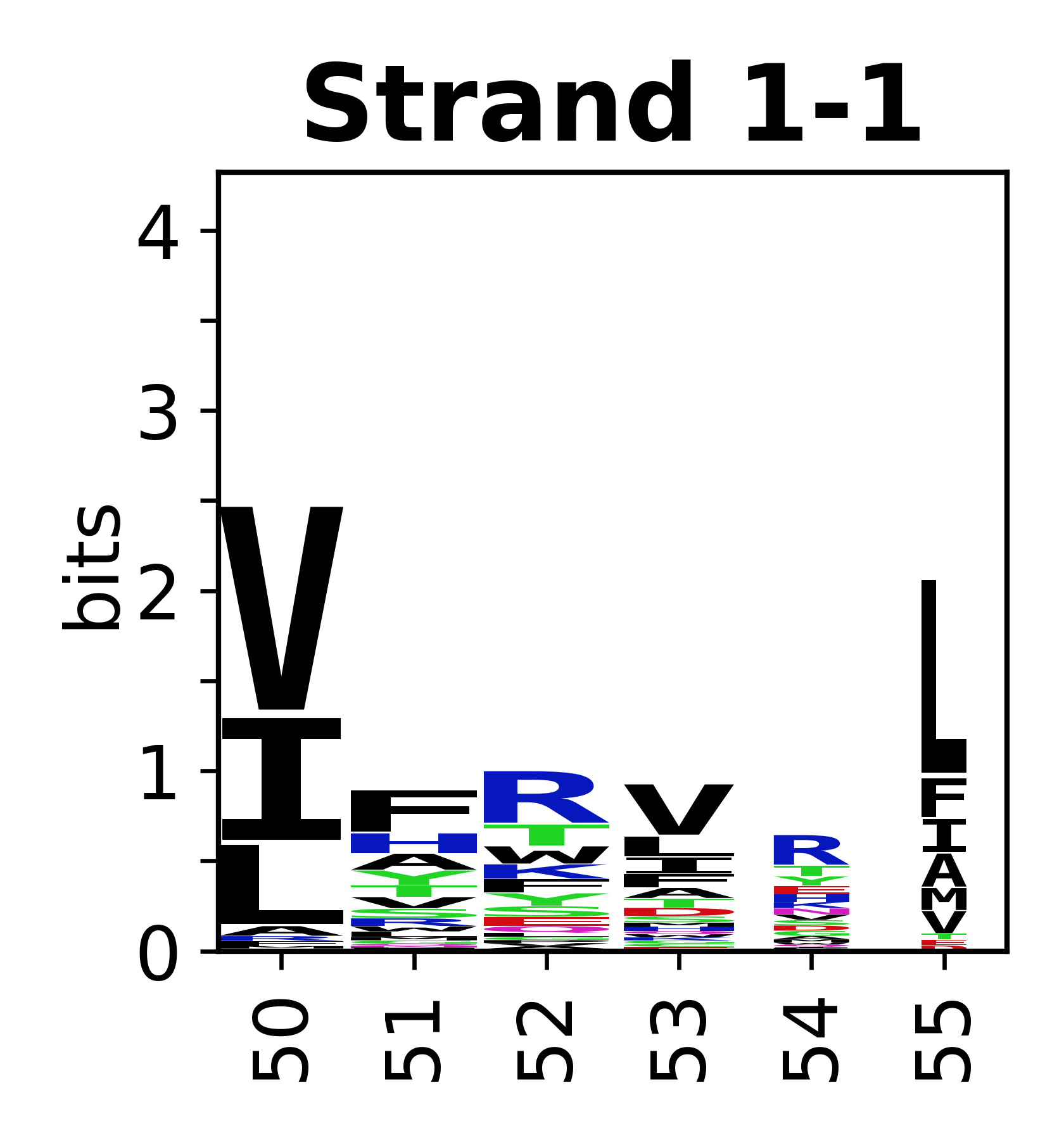
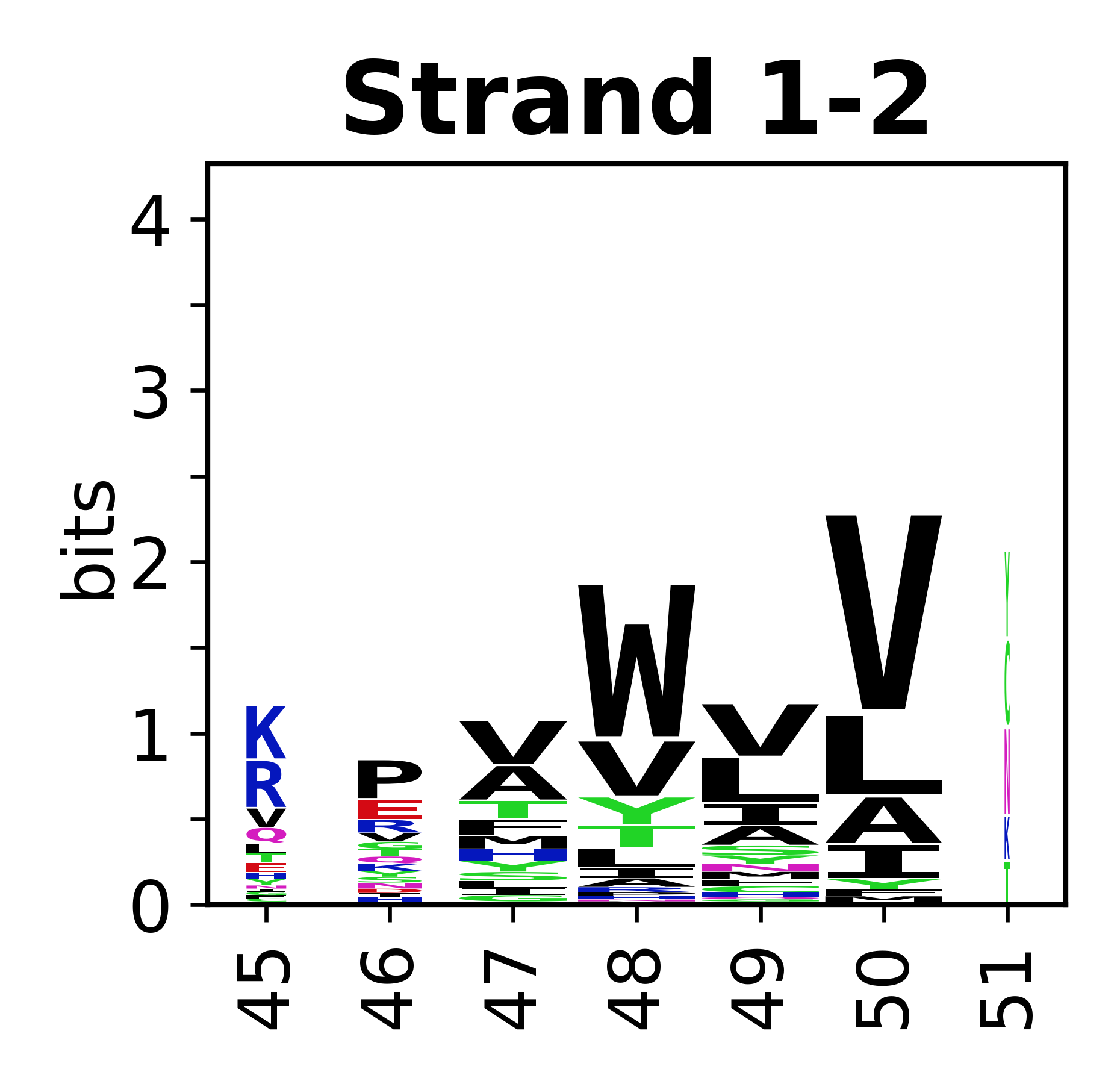
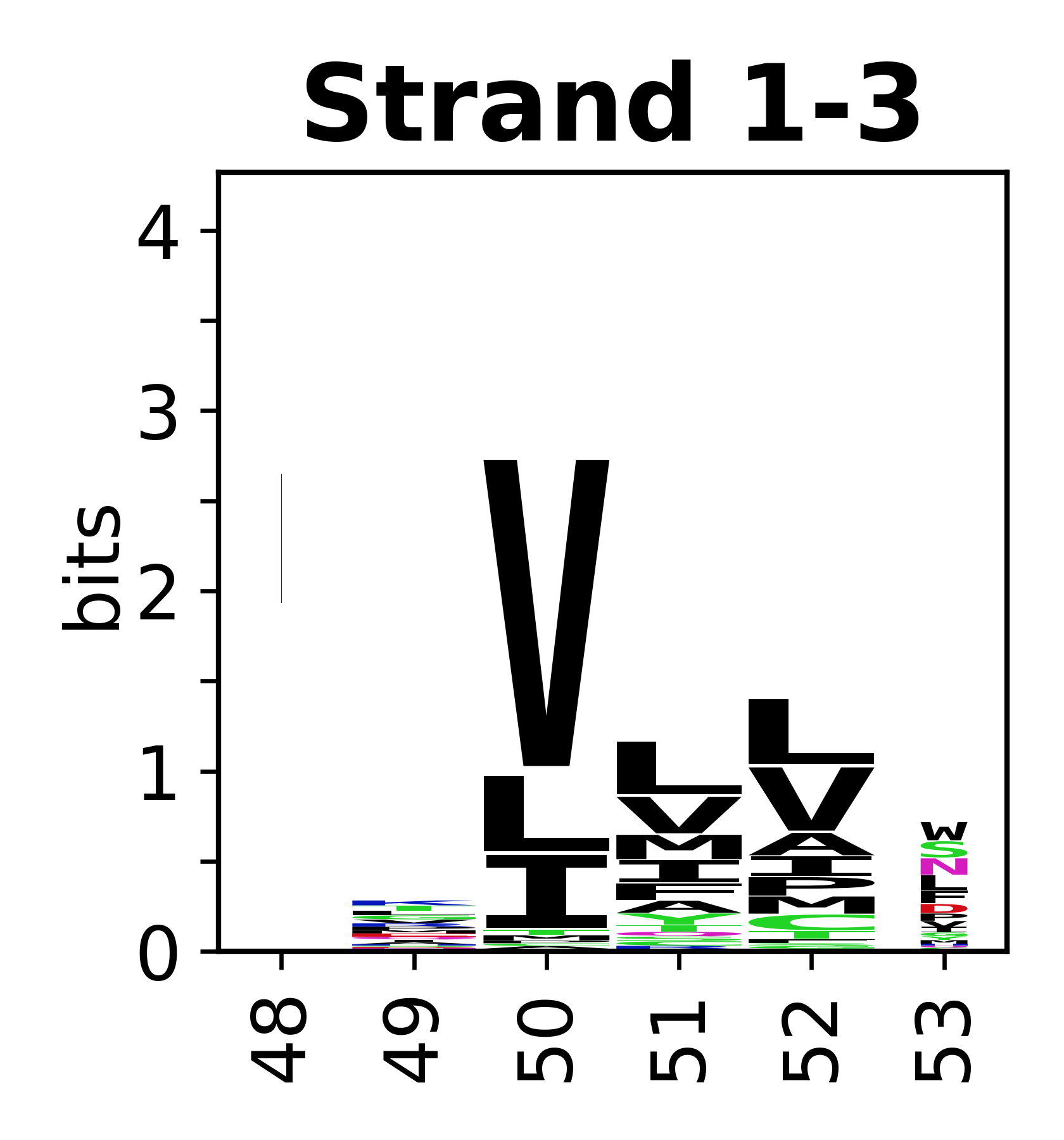
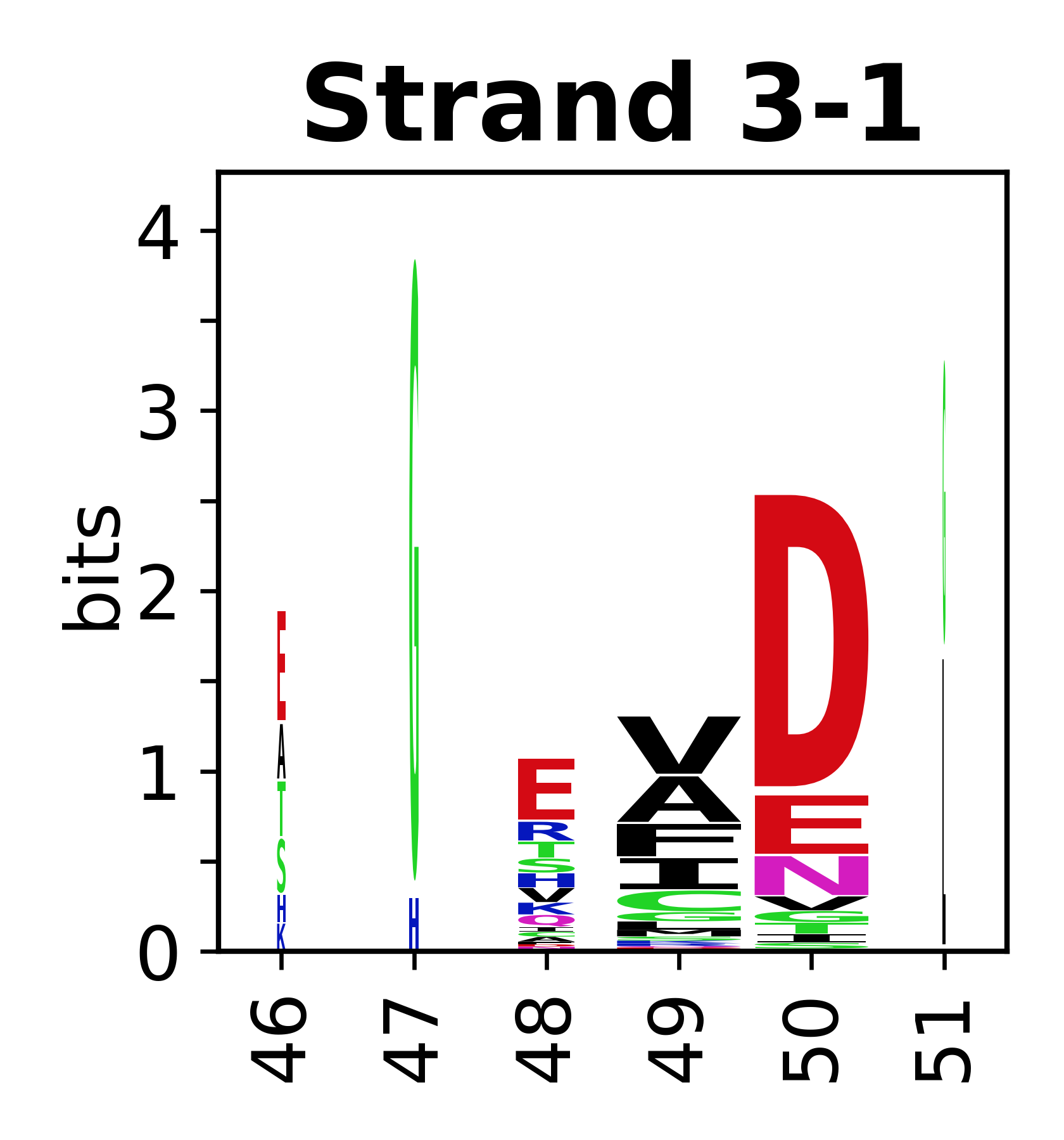
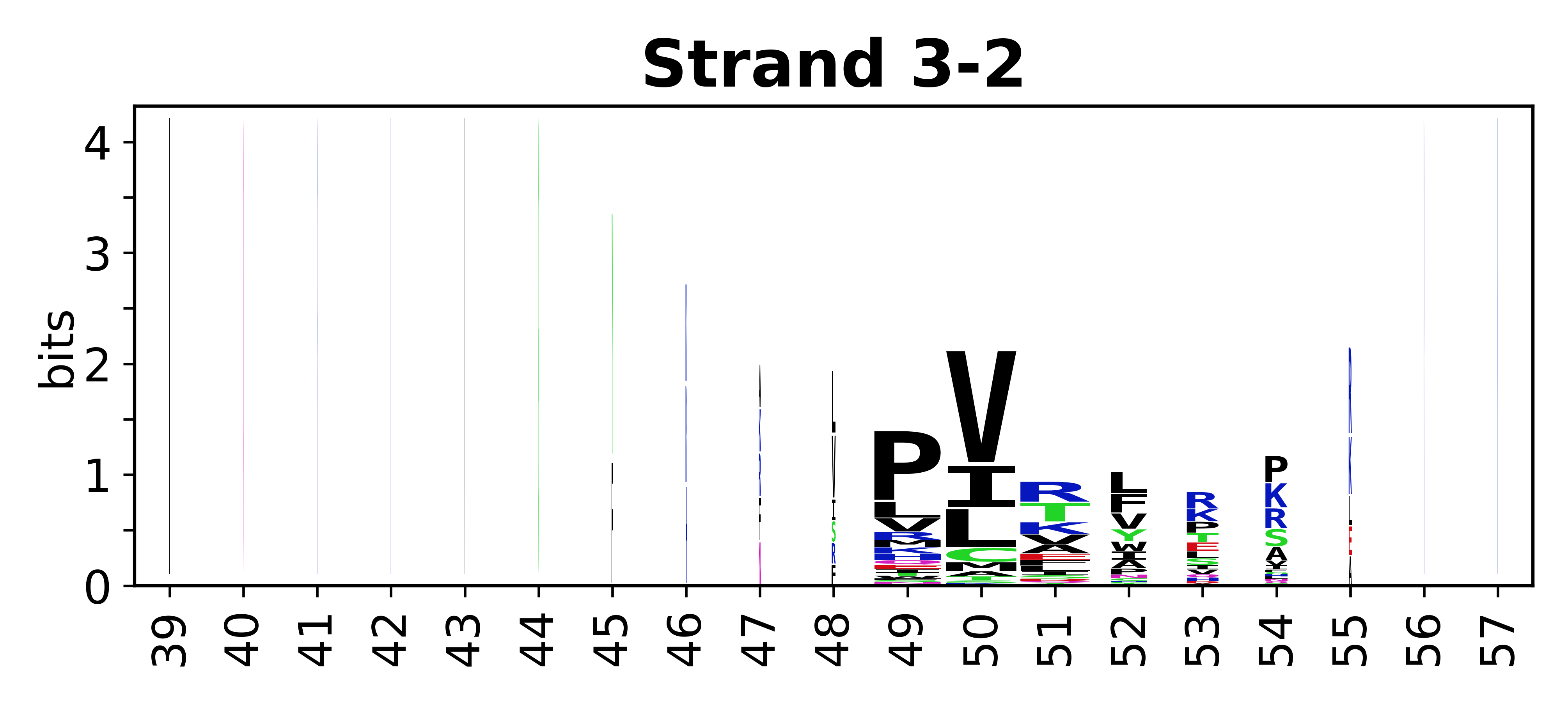
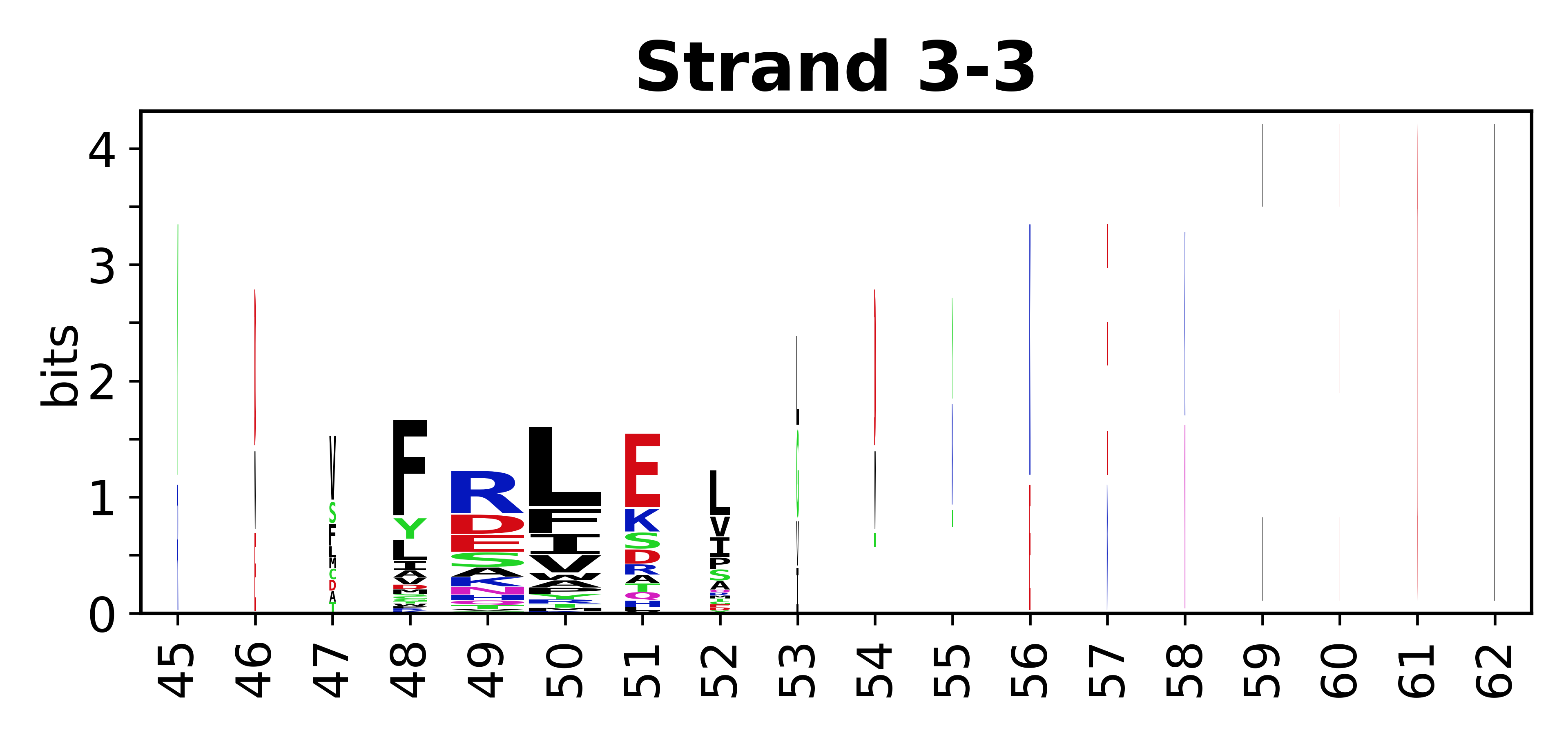
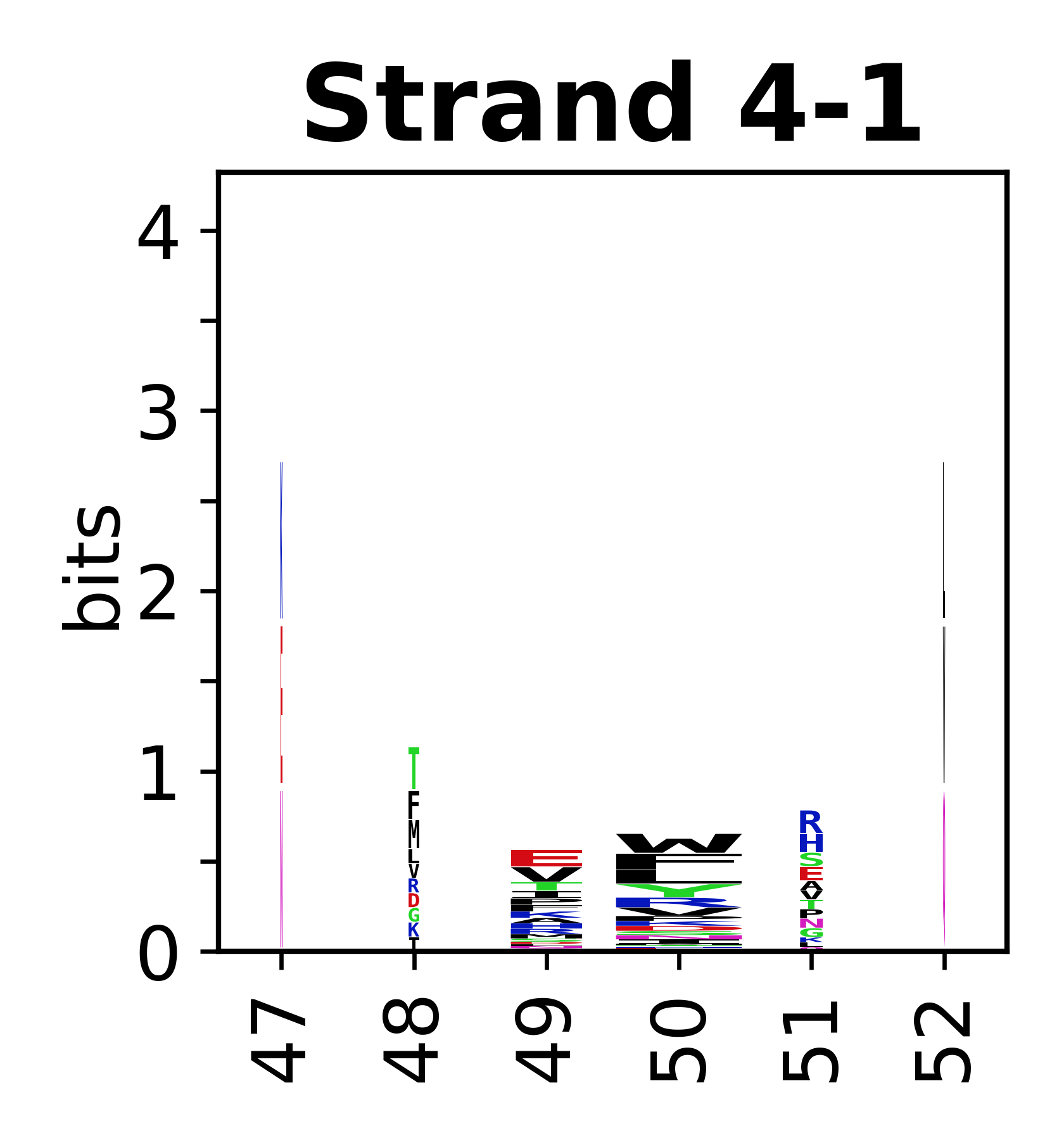
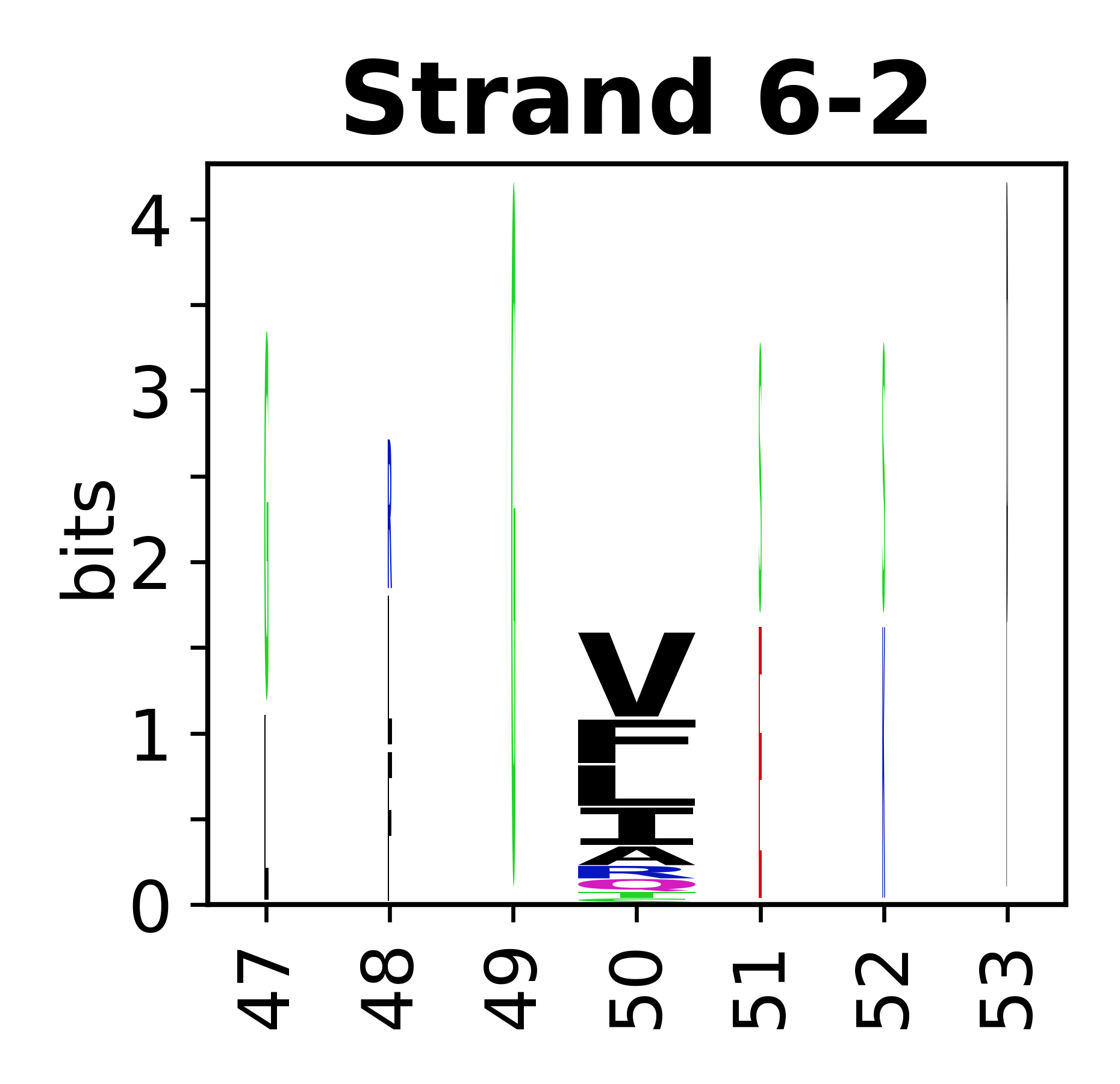
===Sequence of SSEs=== | |||
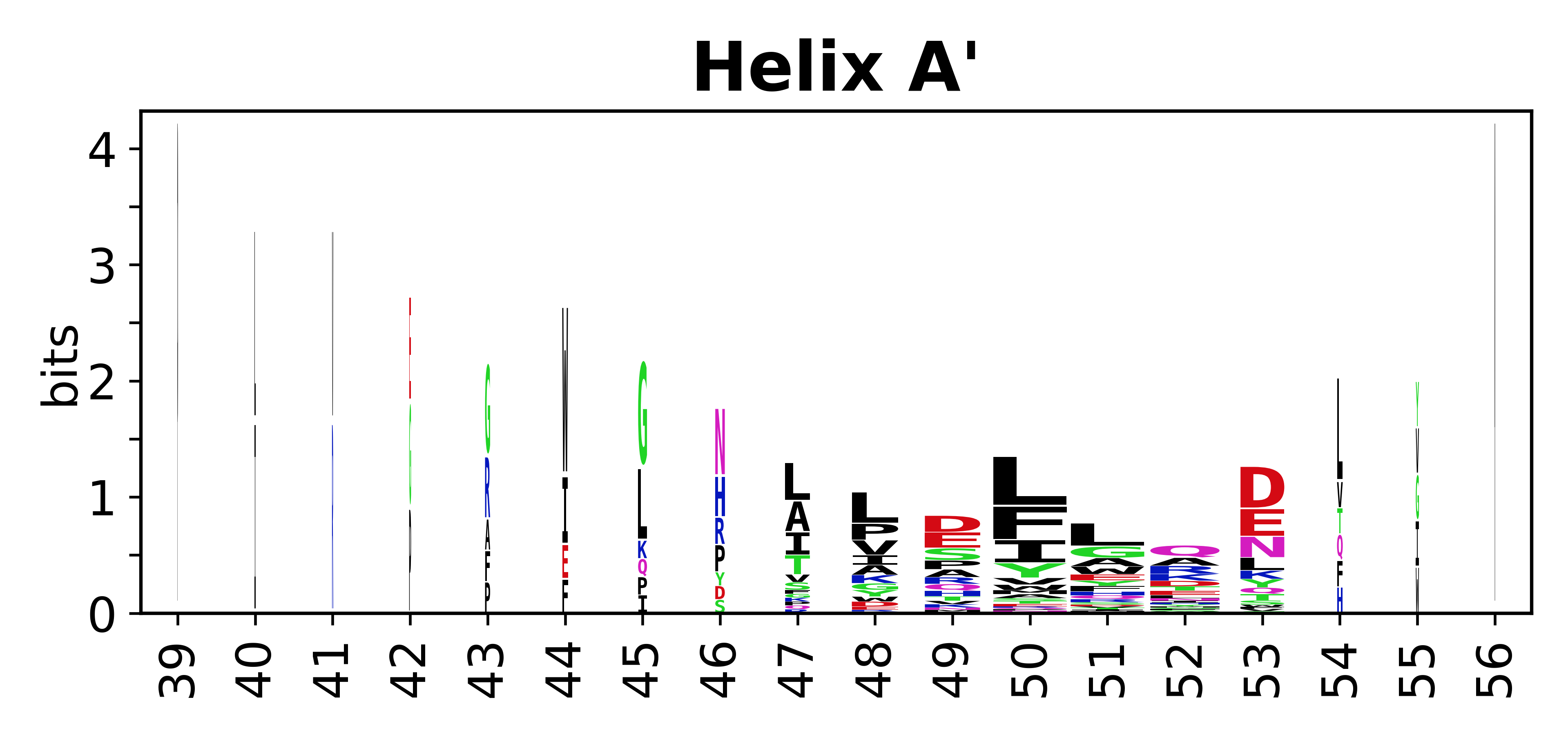
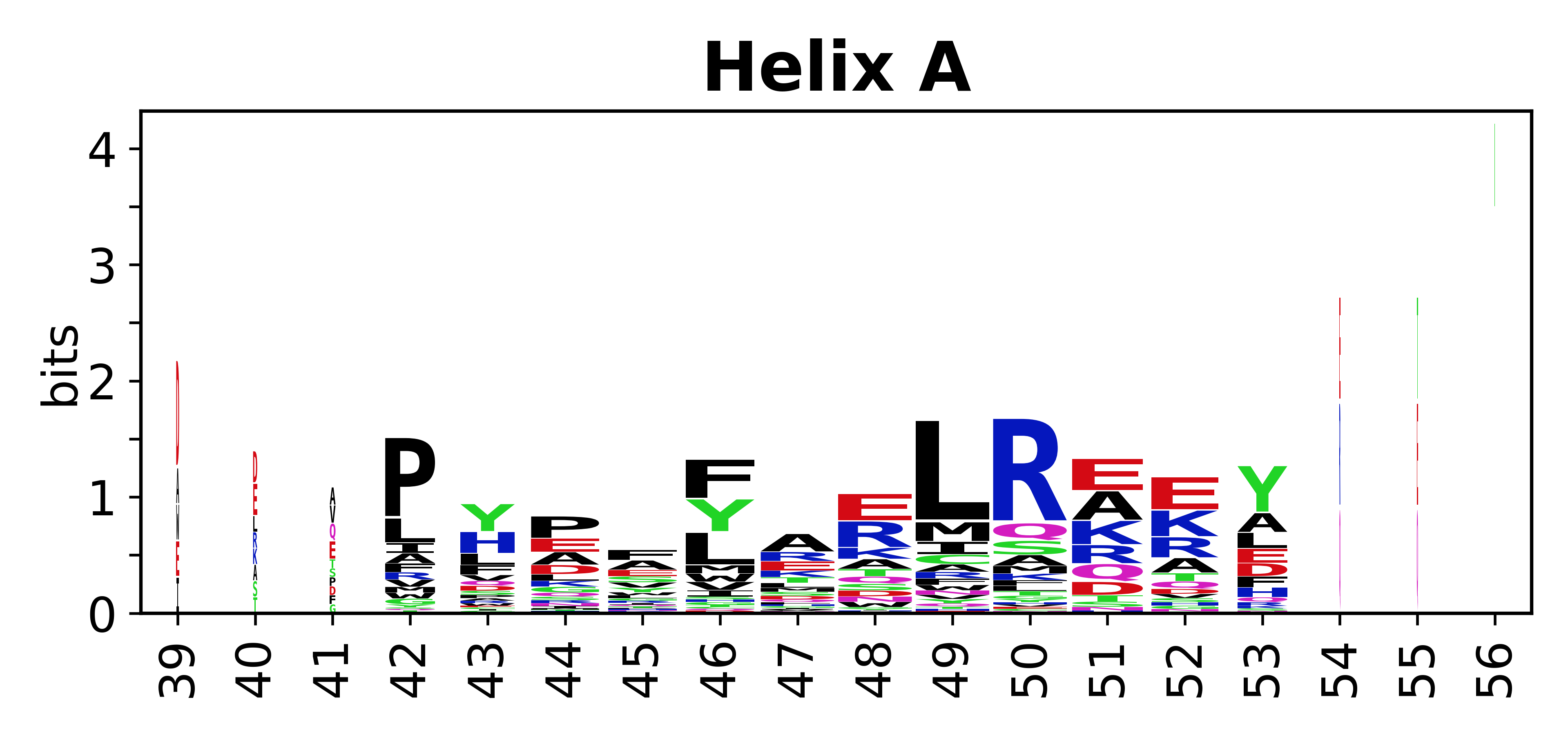
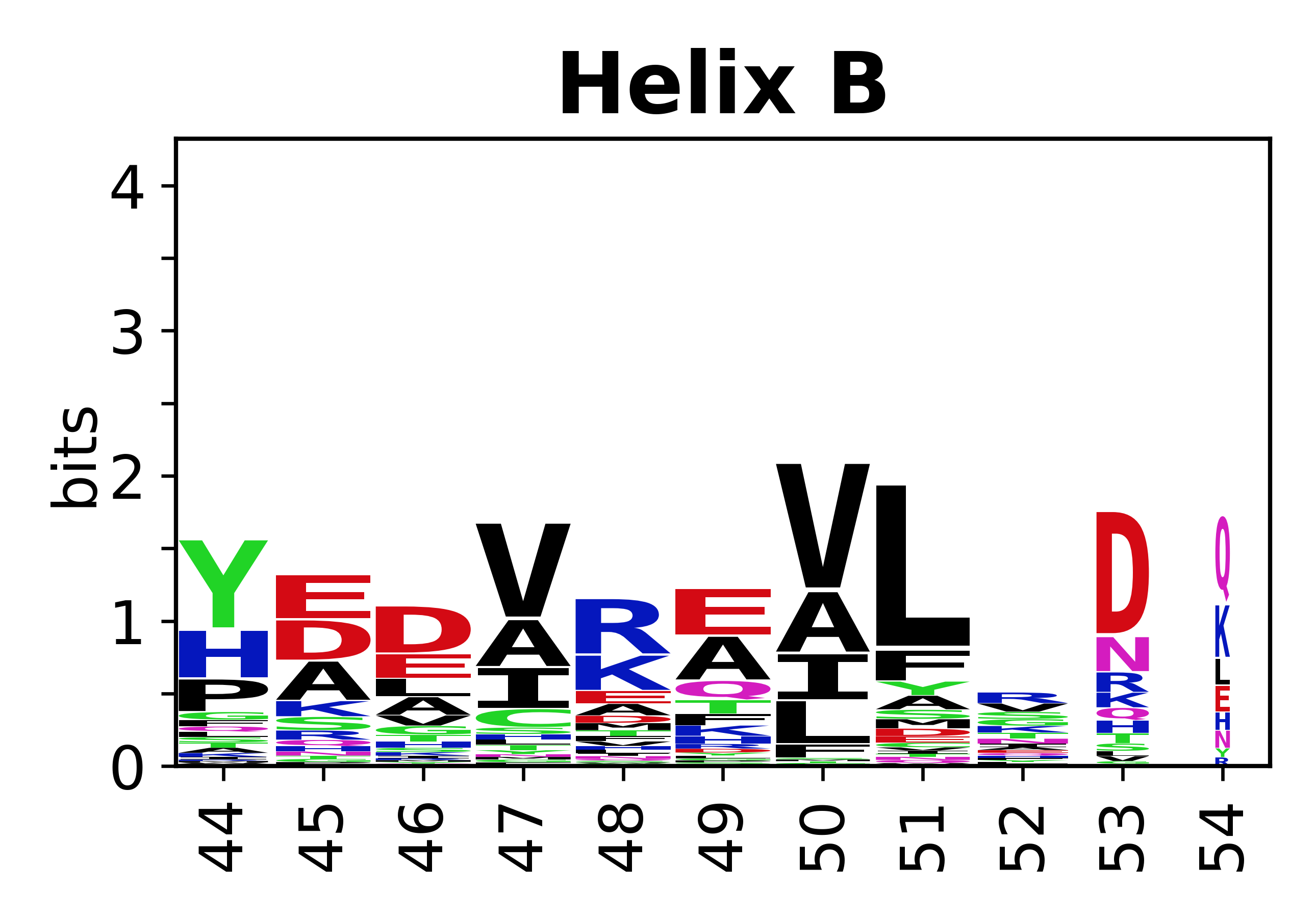
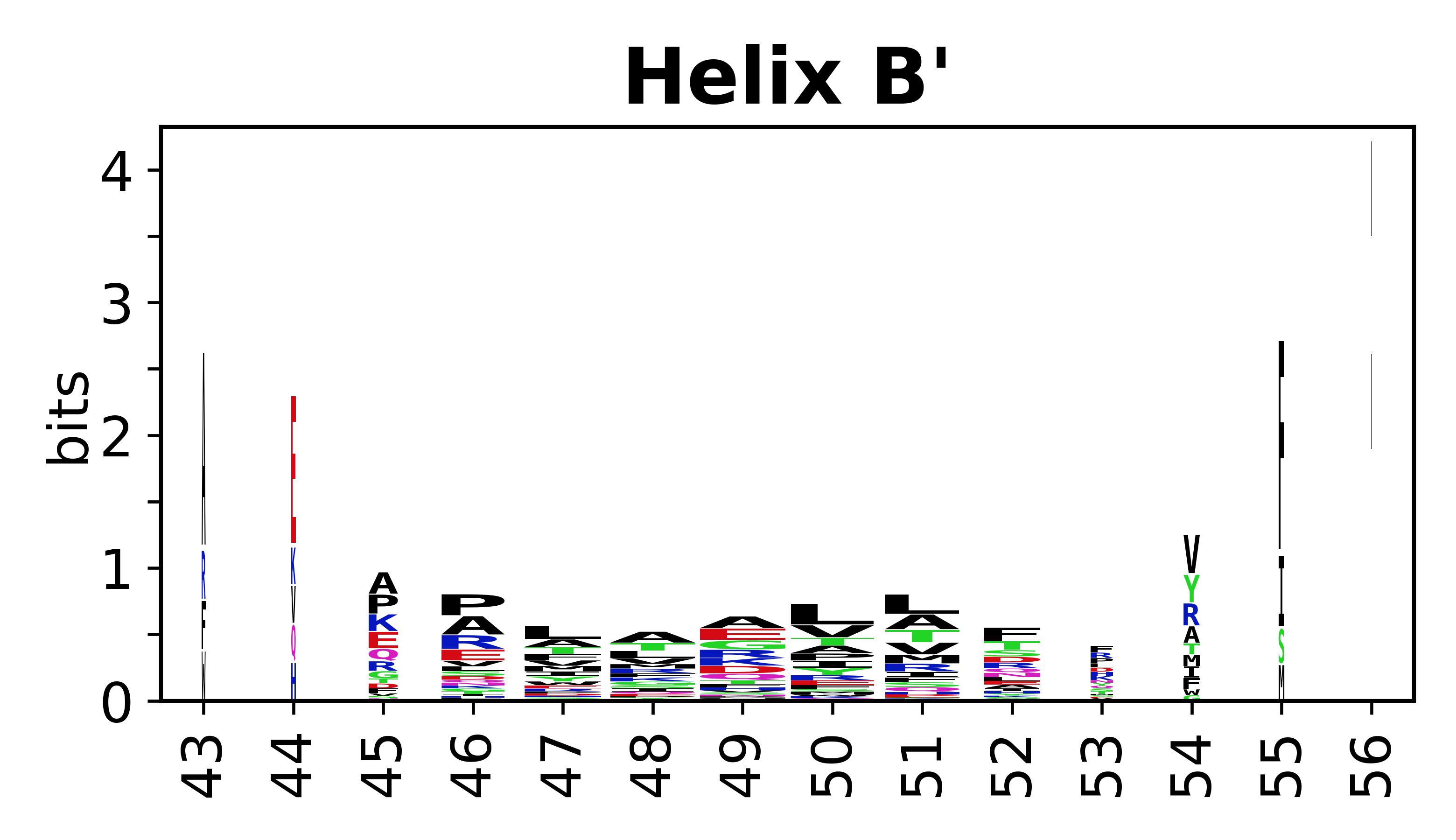
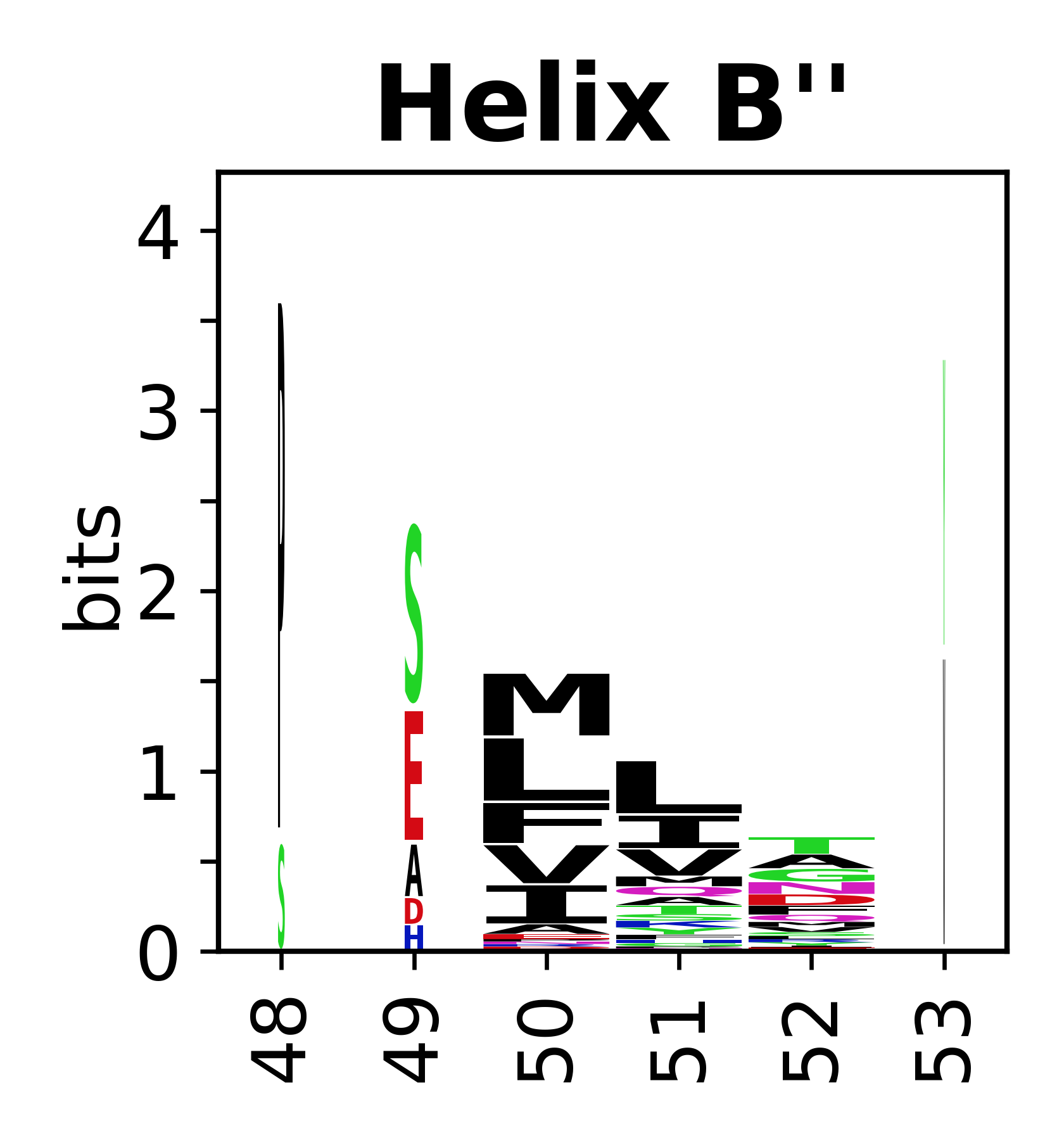
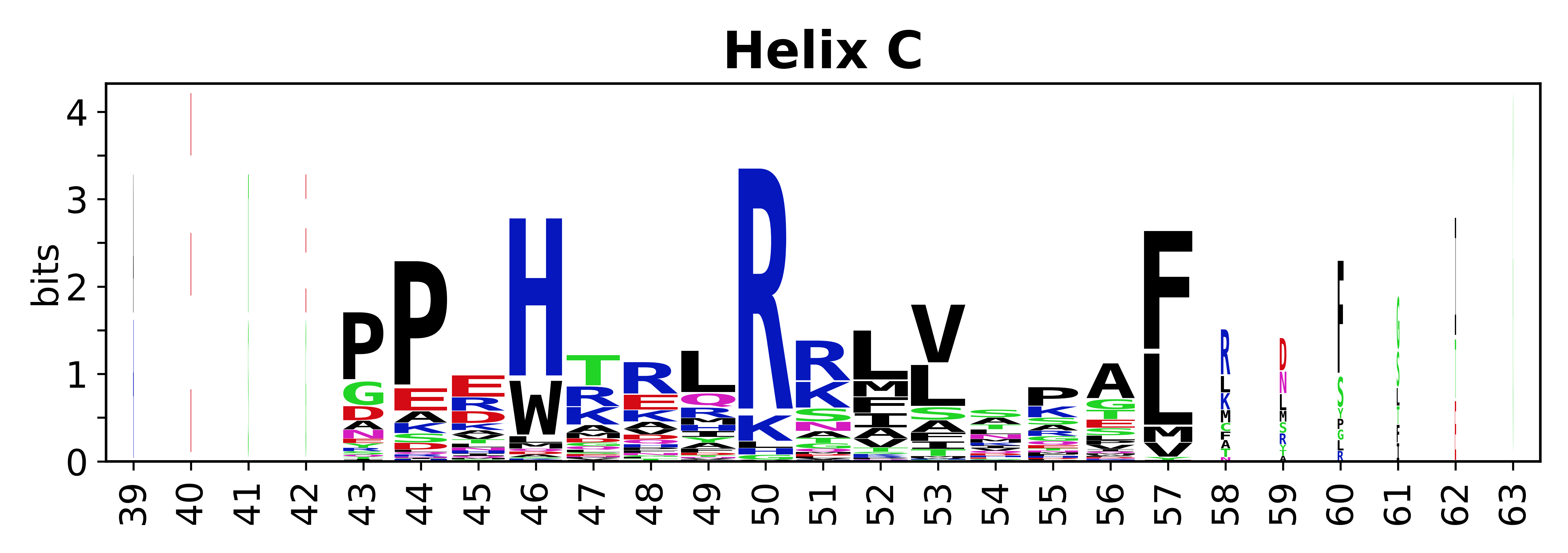
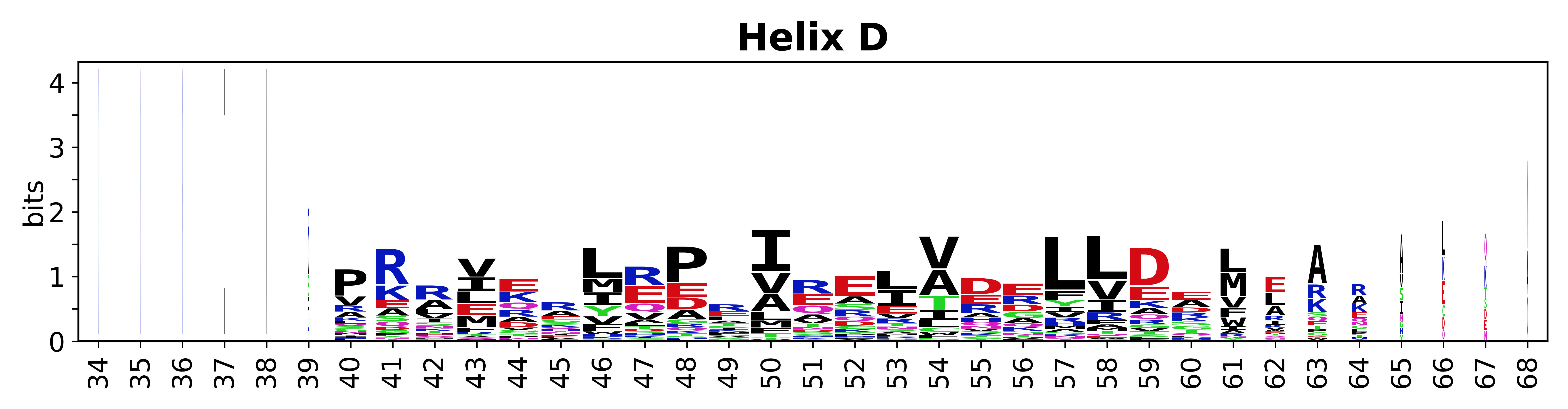
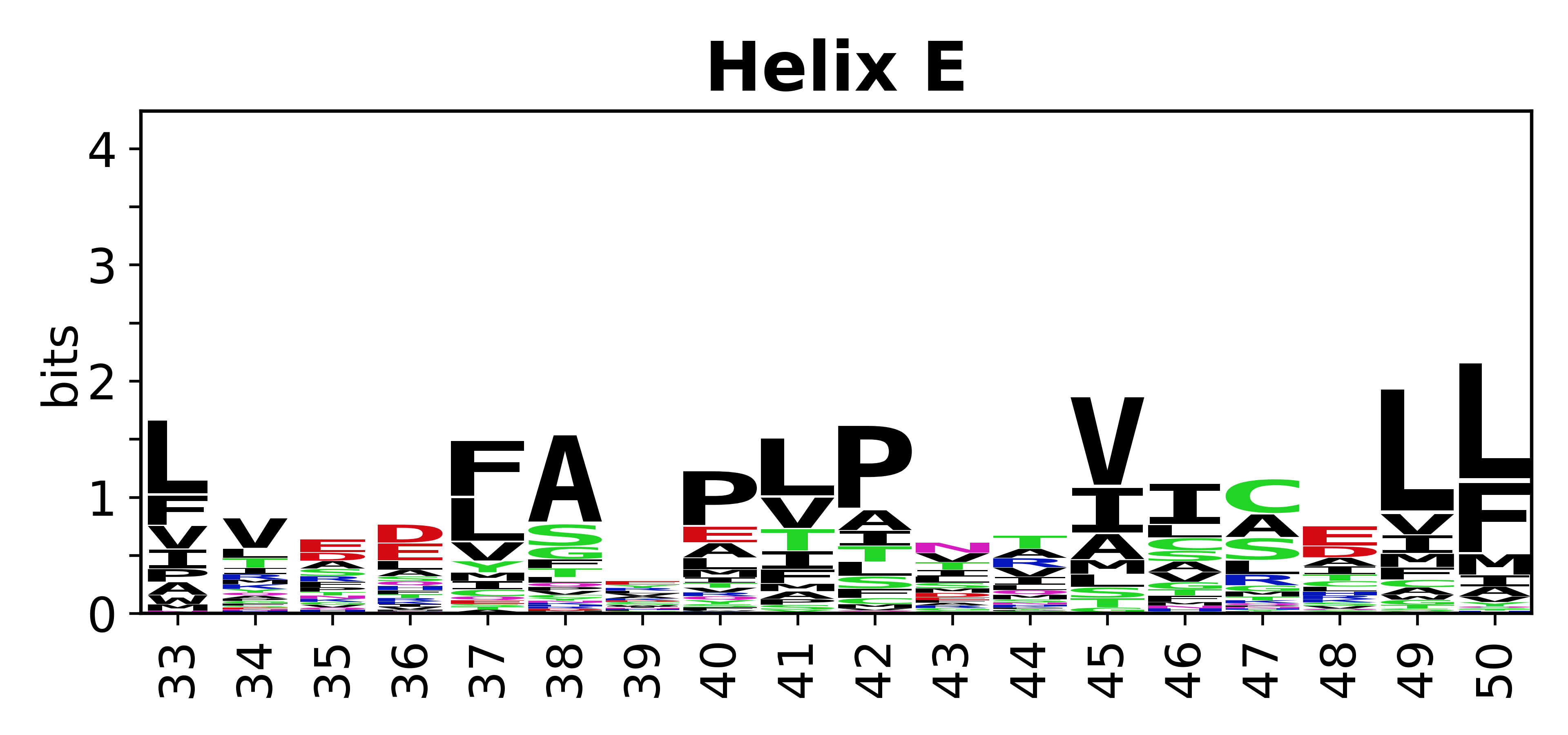
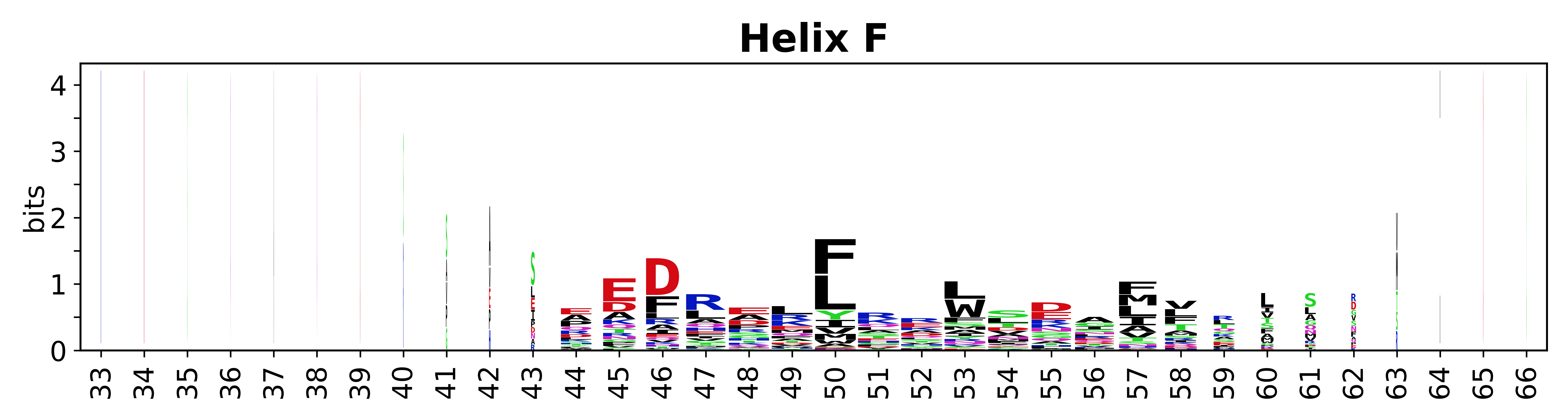
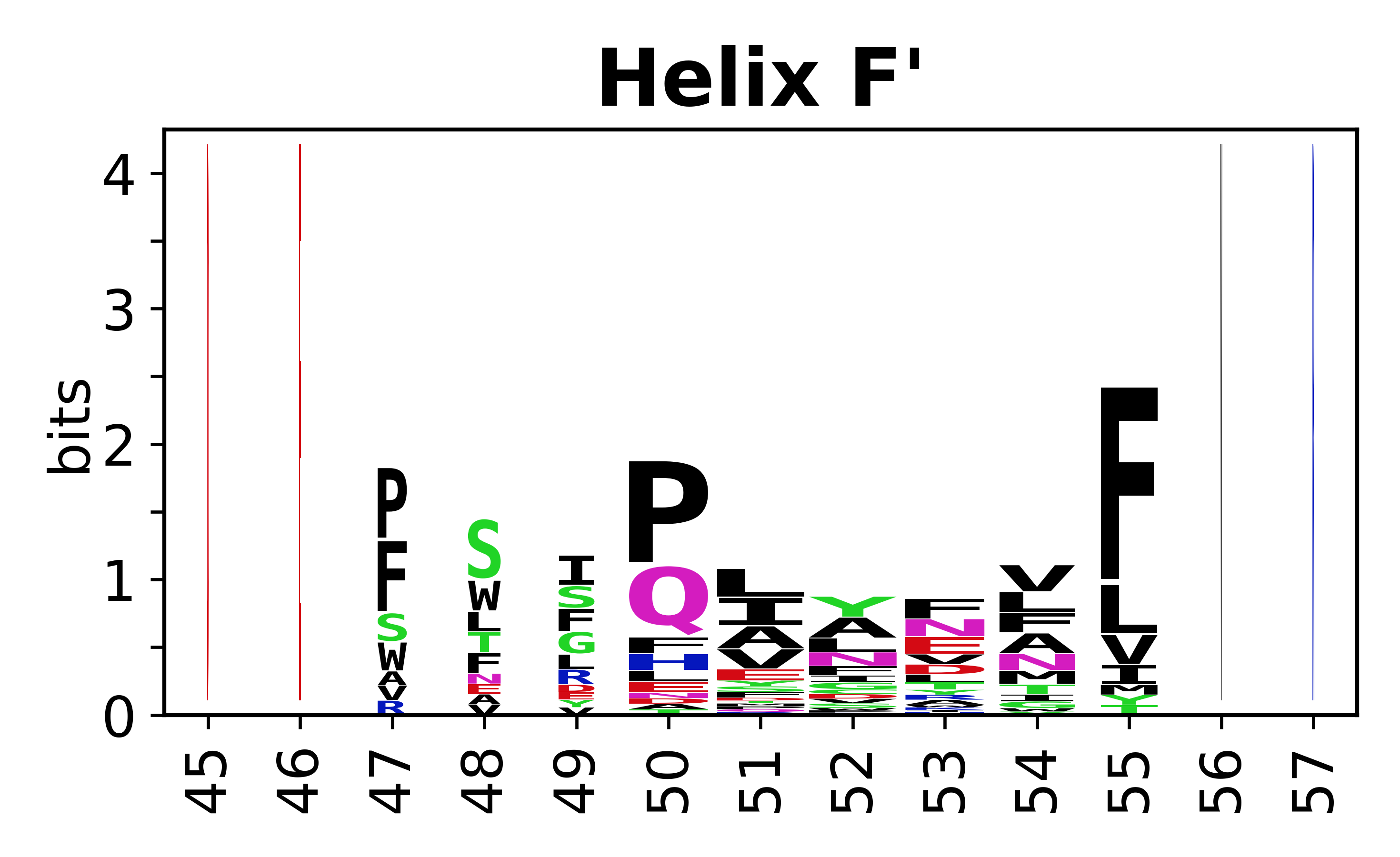
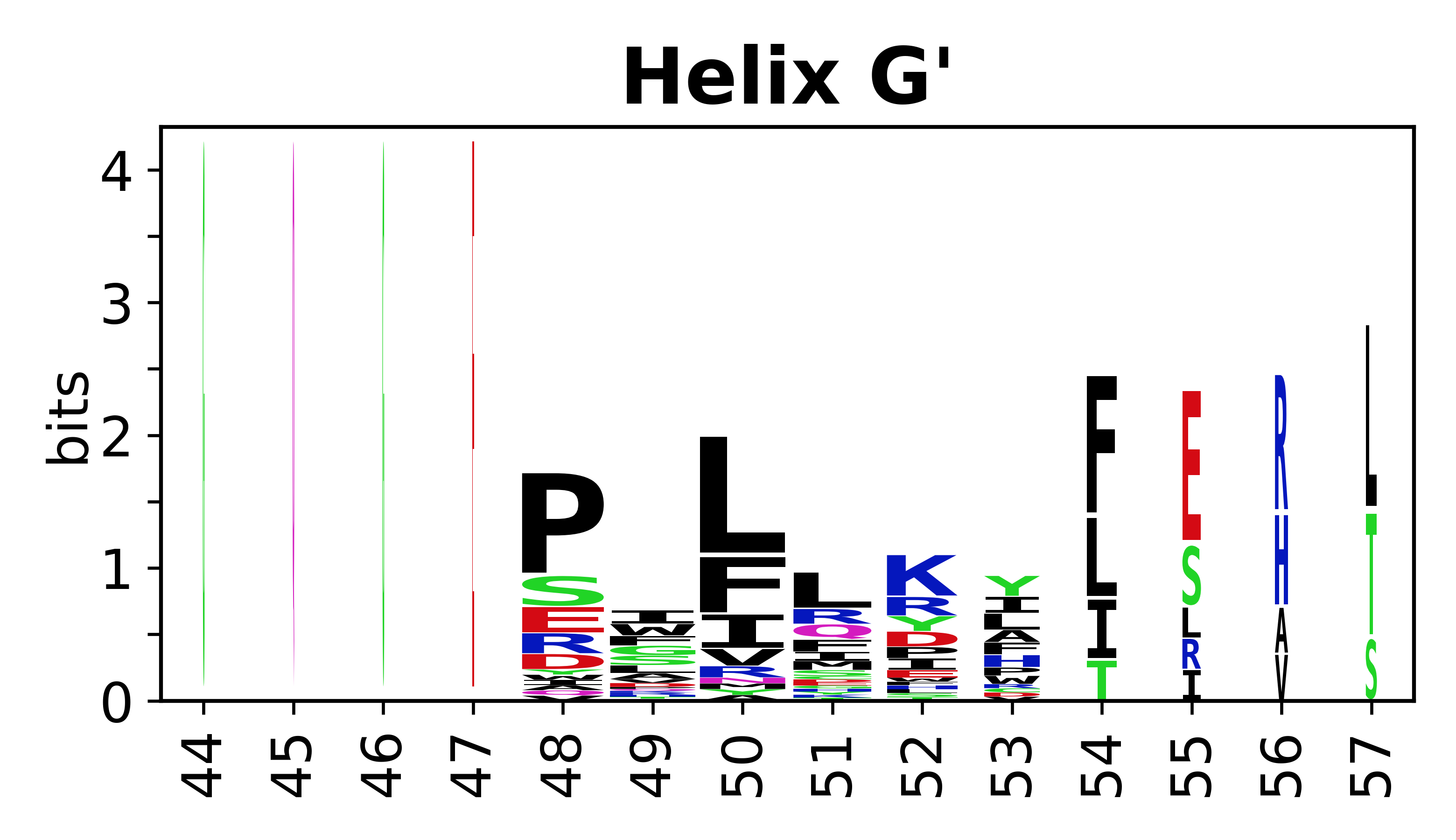
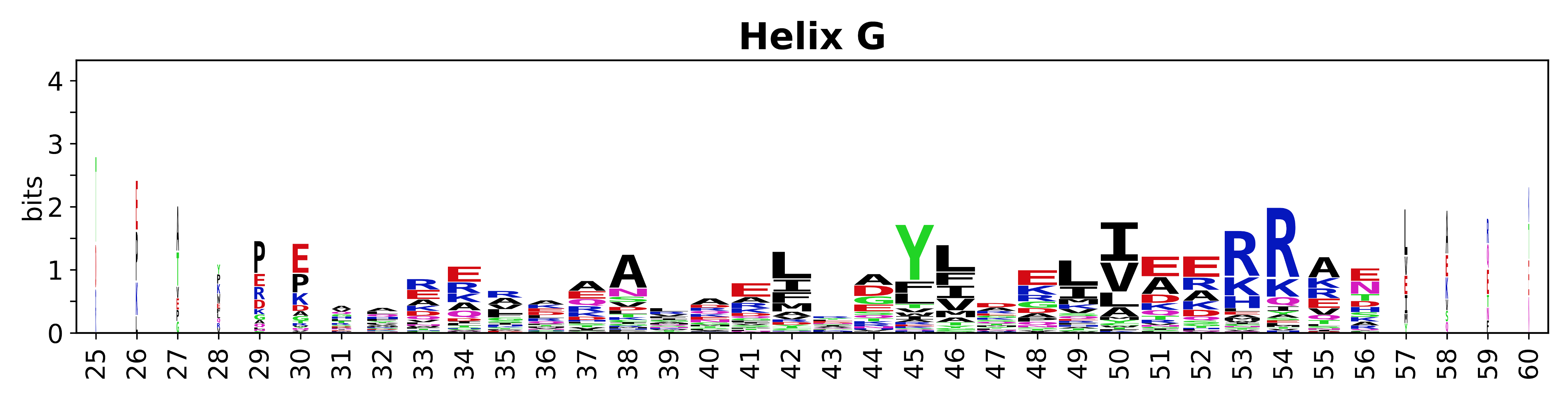
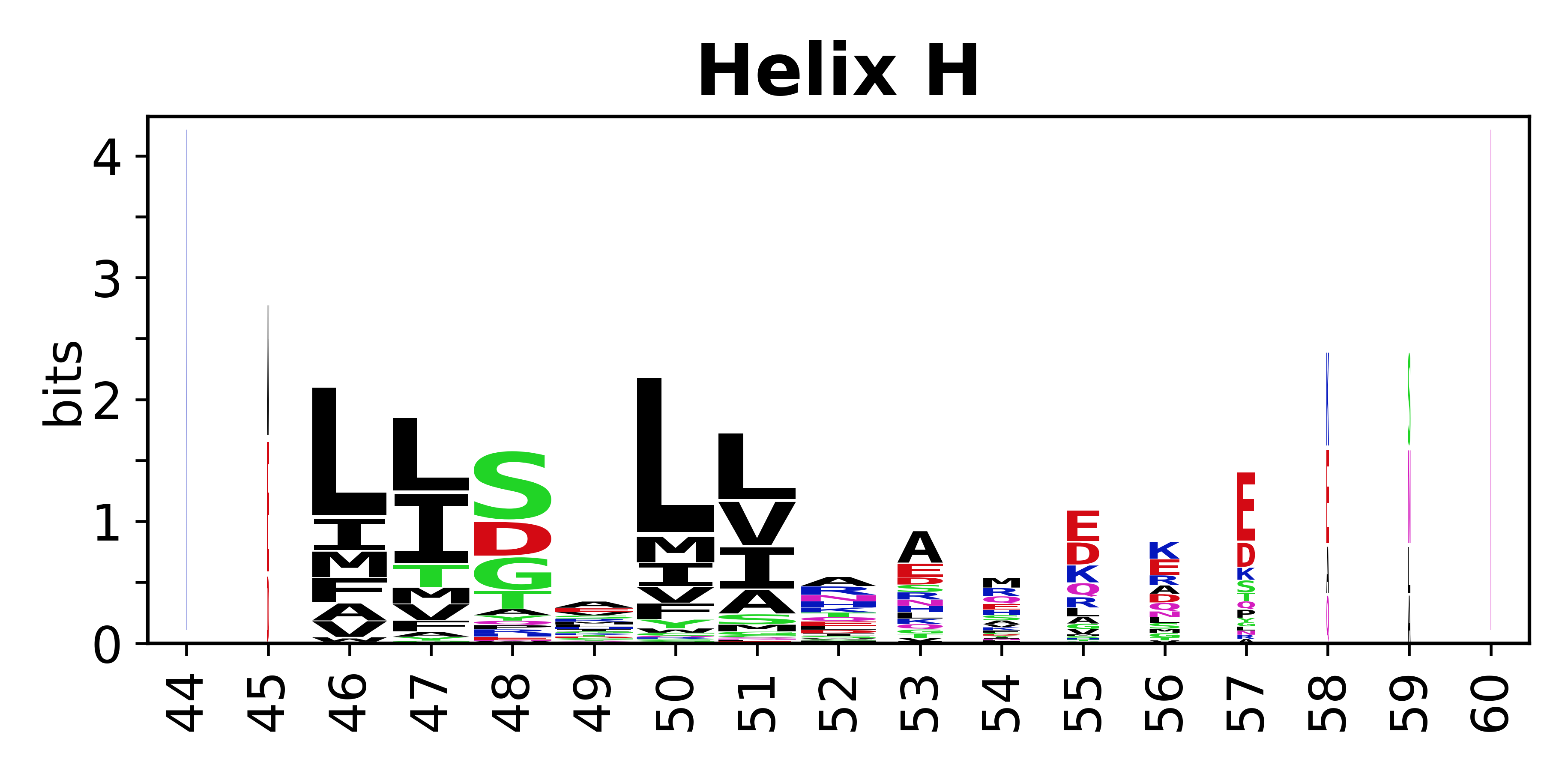
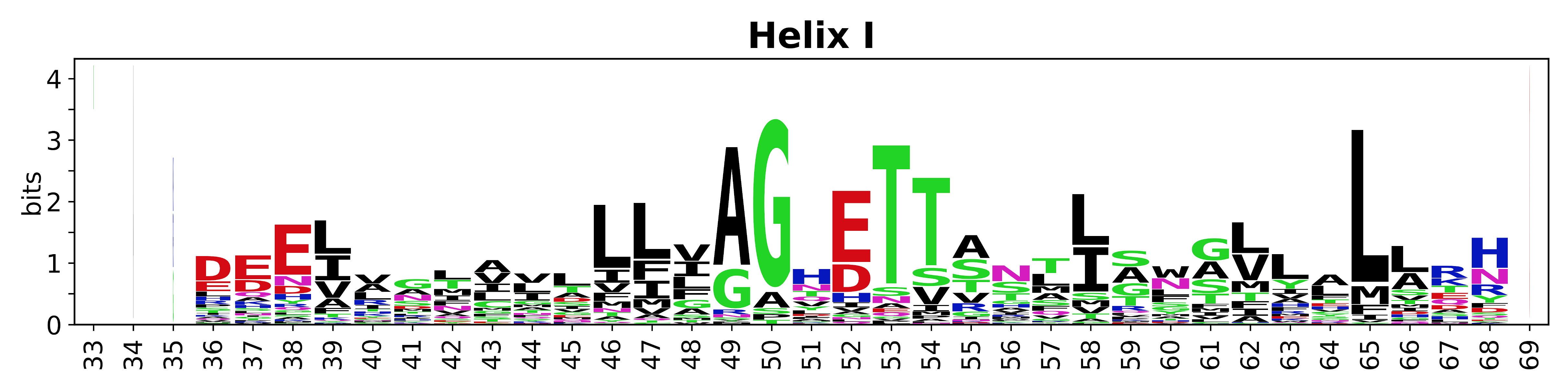
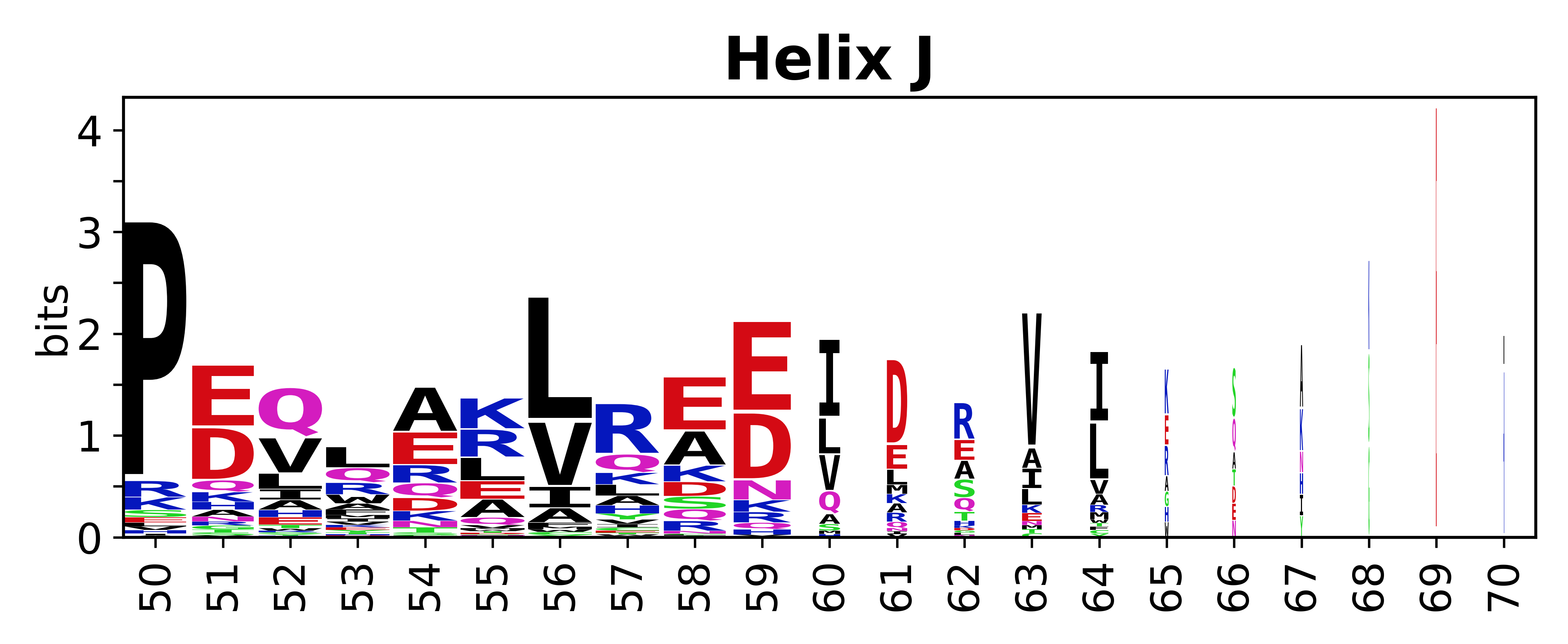
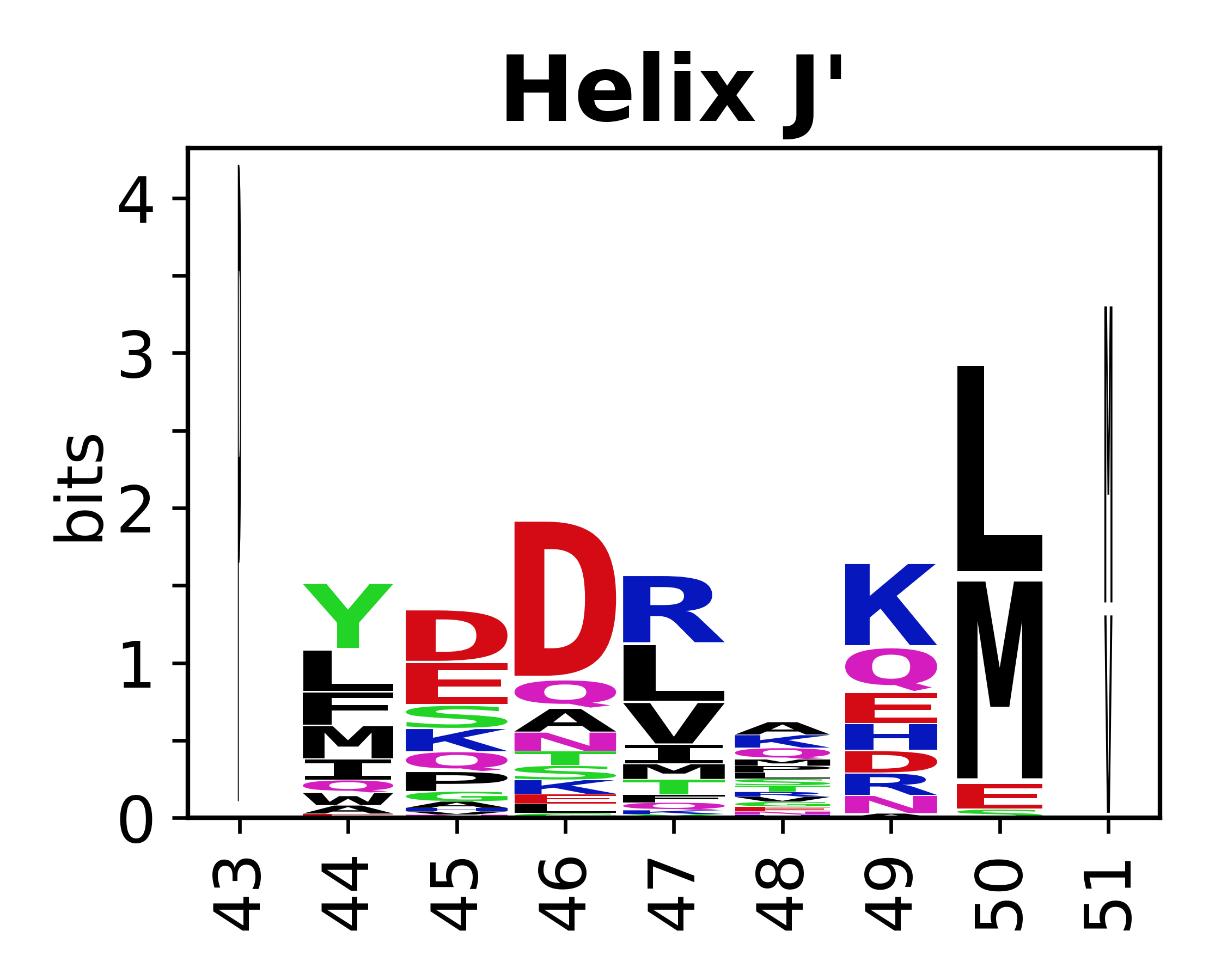
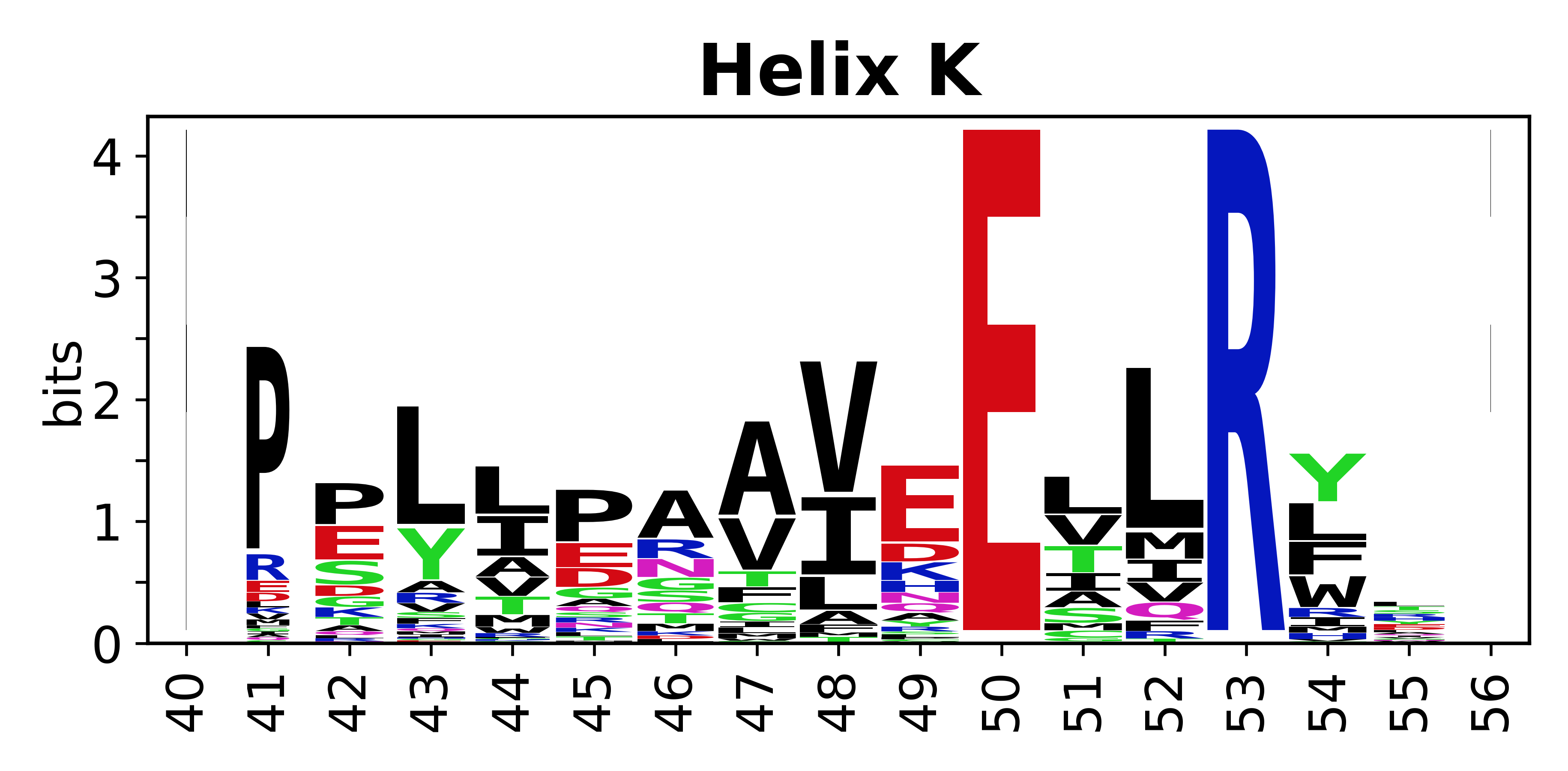
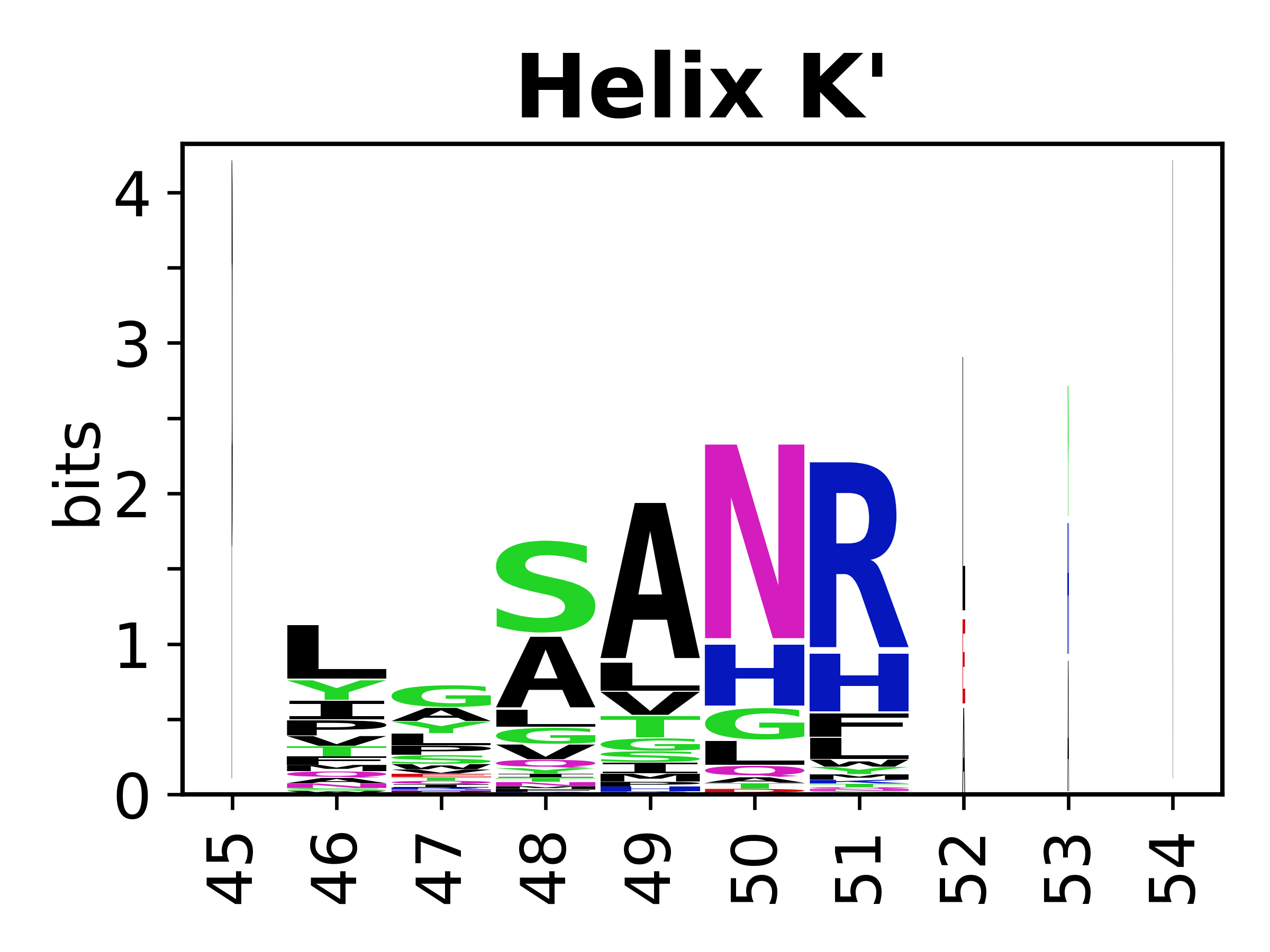
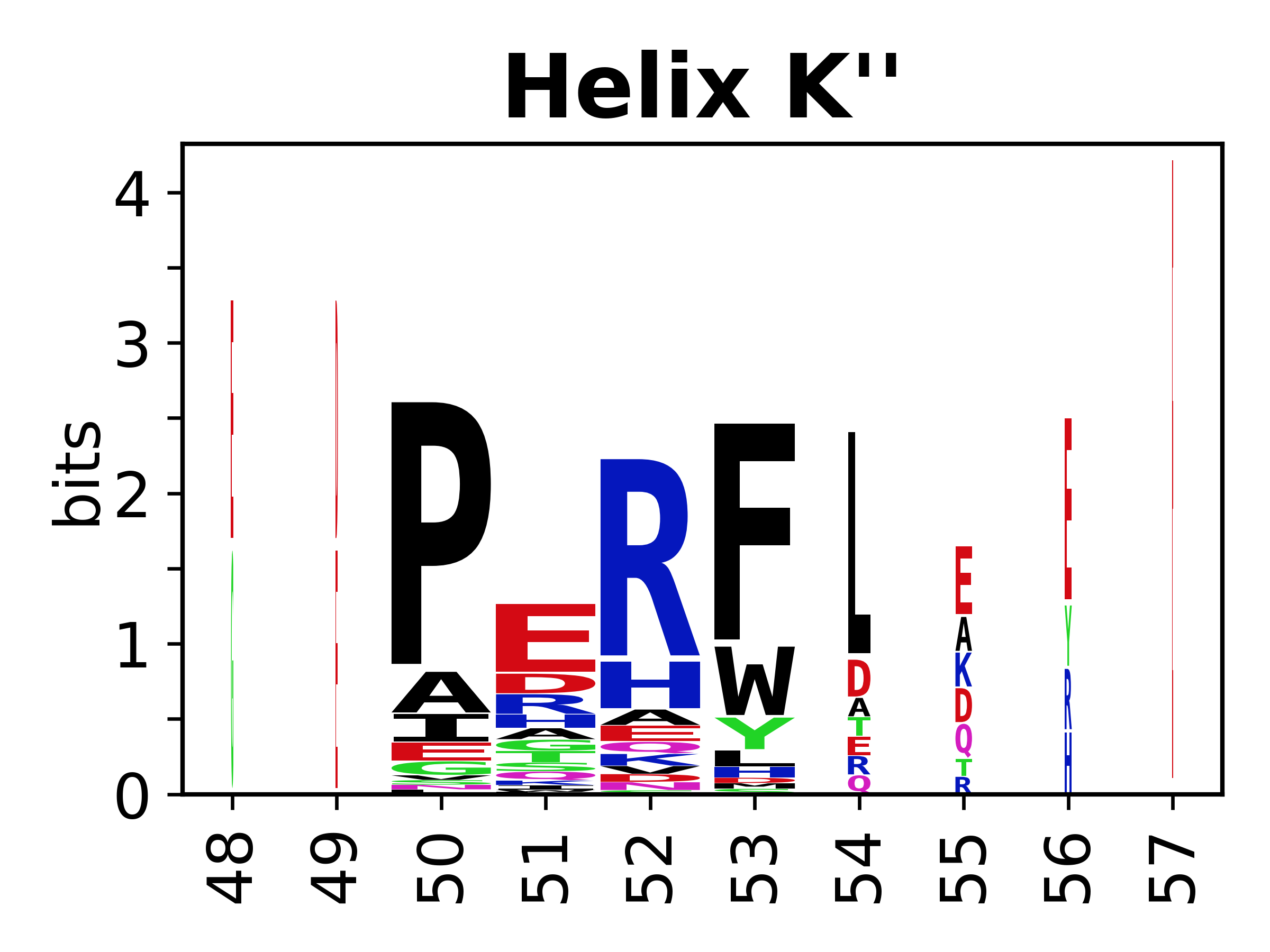
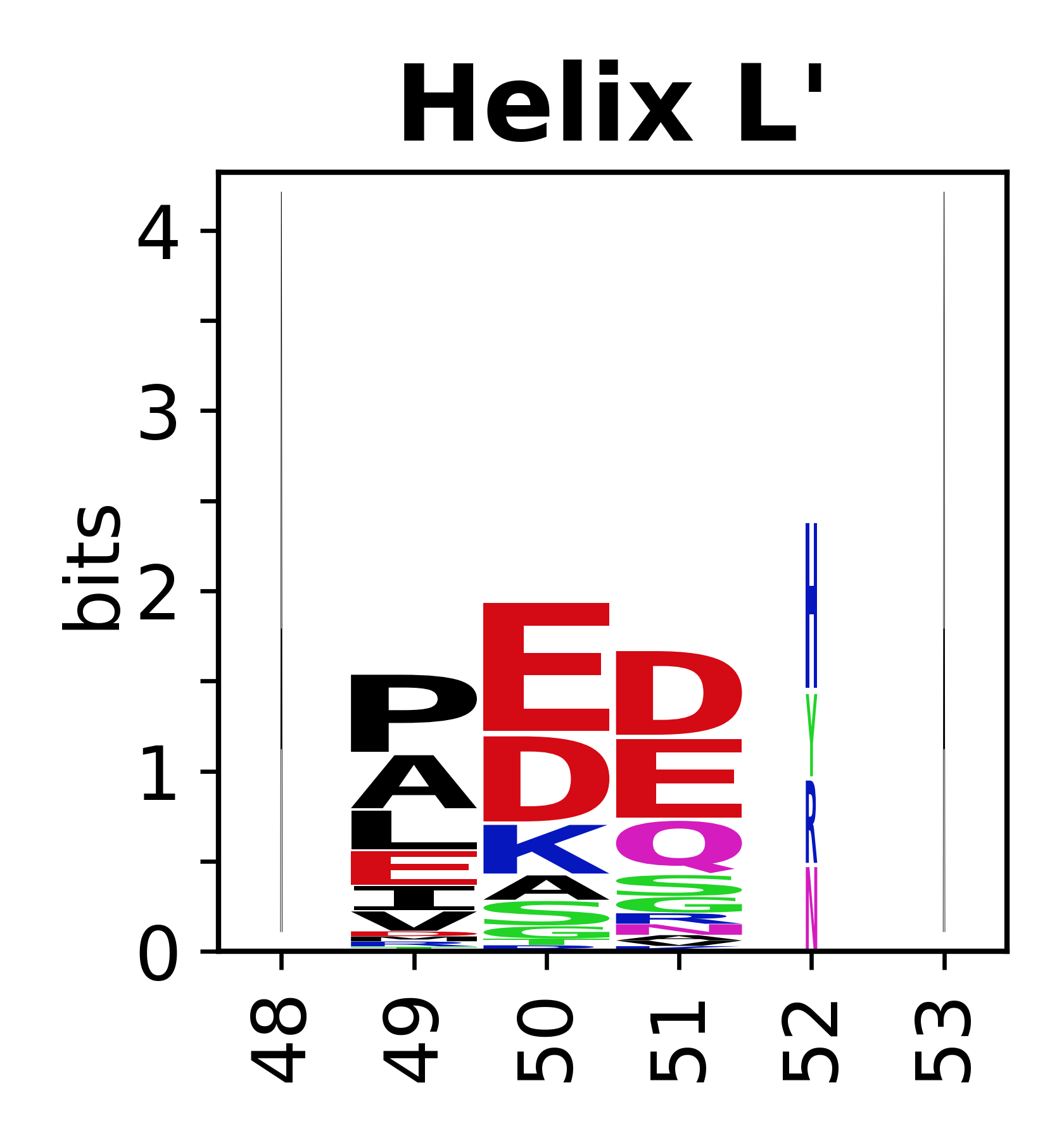
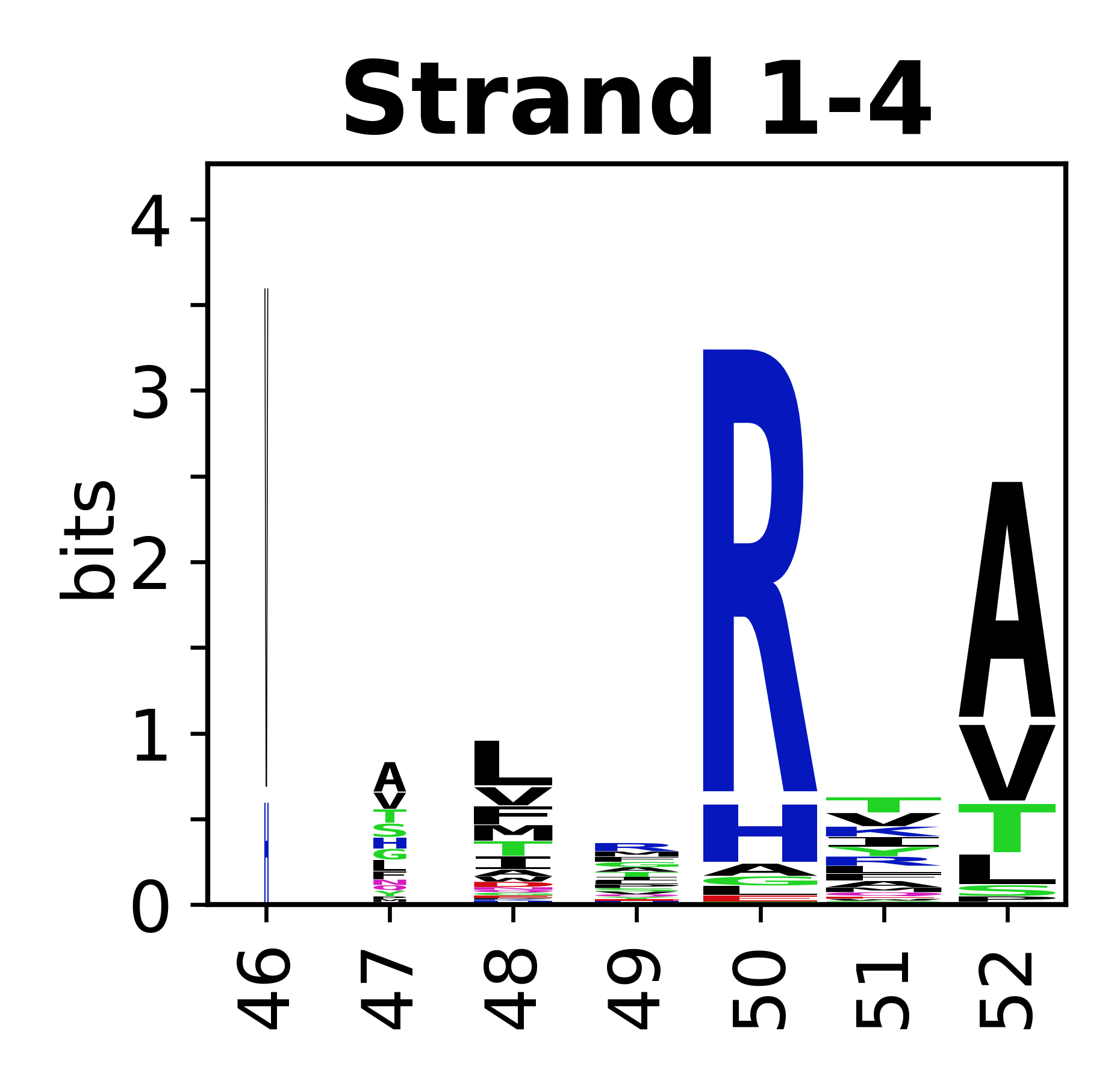
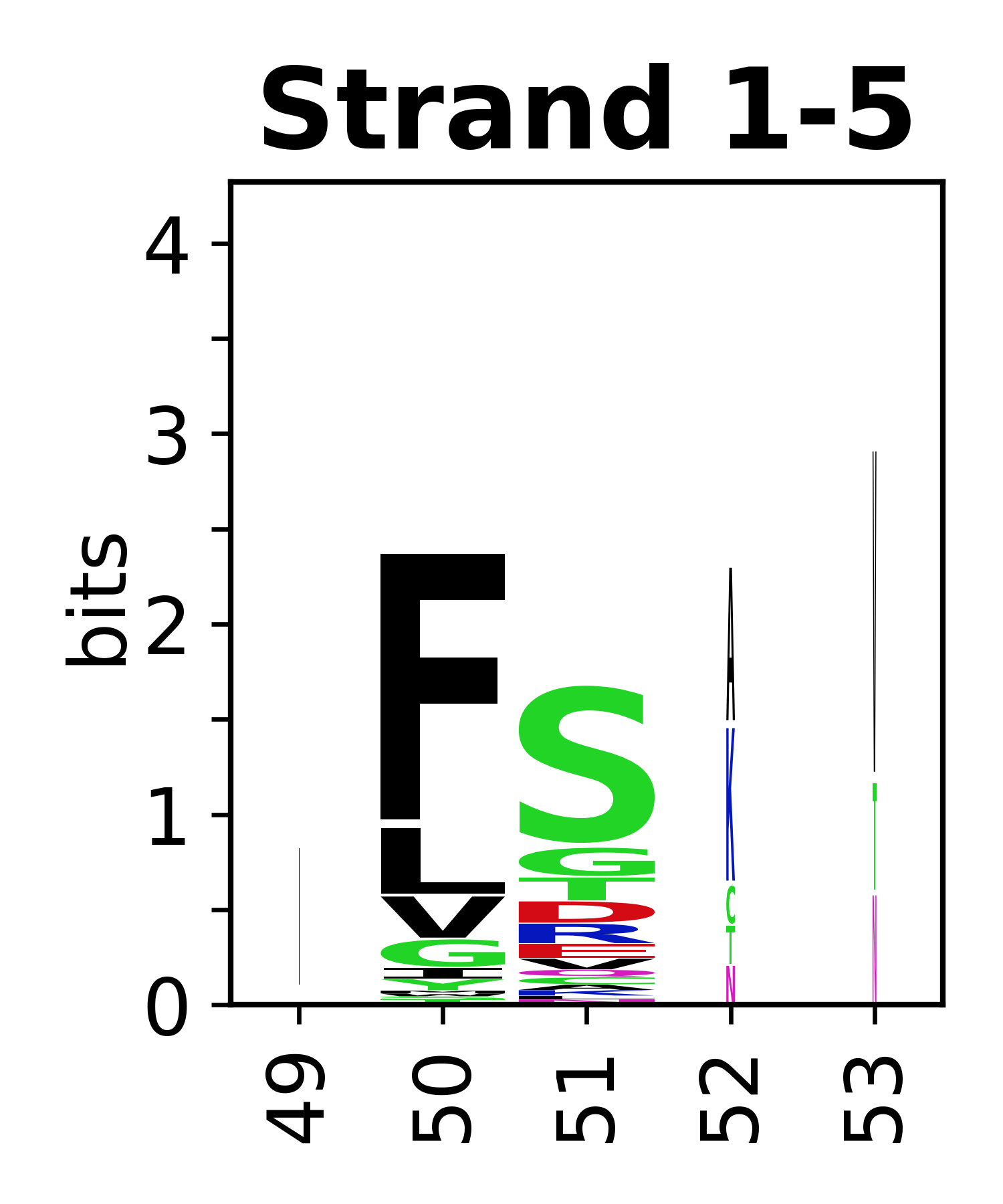
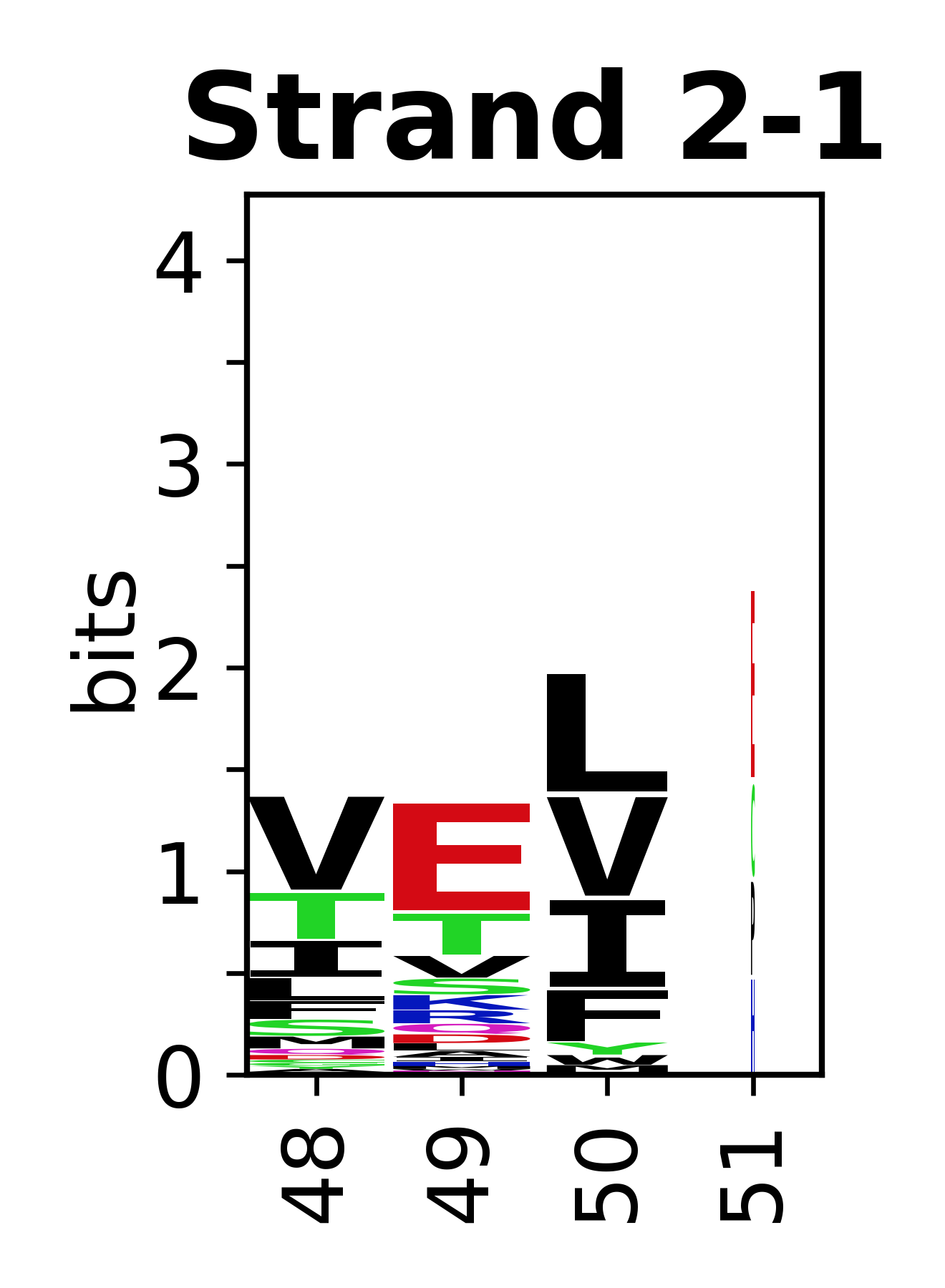
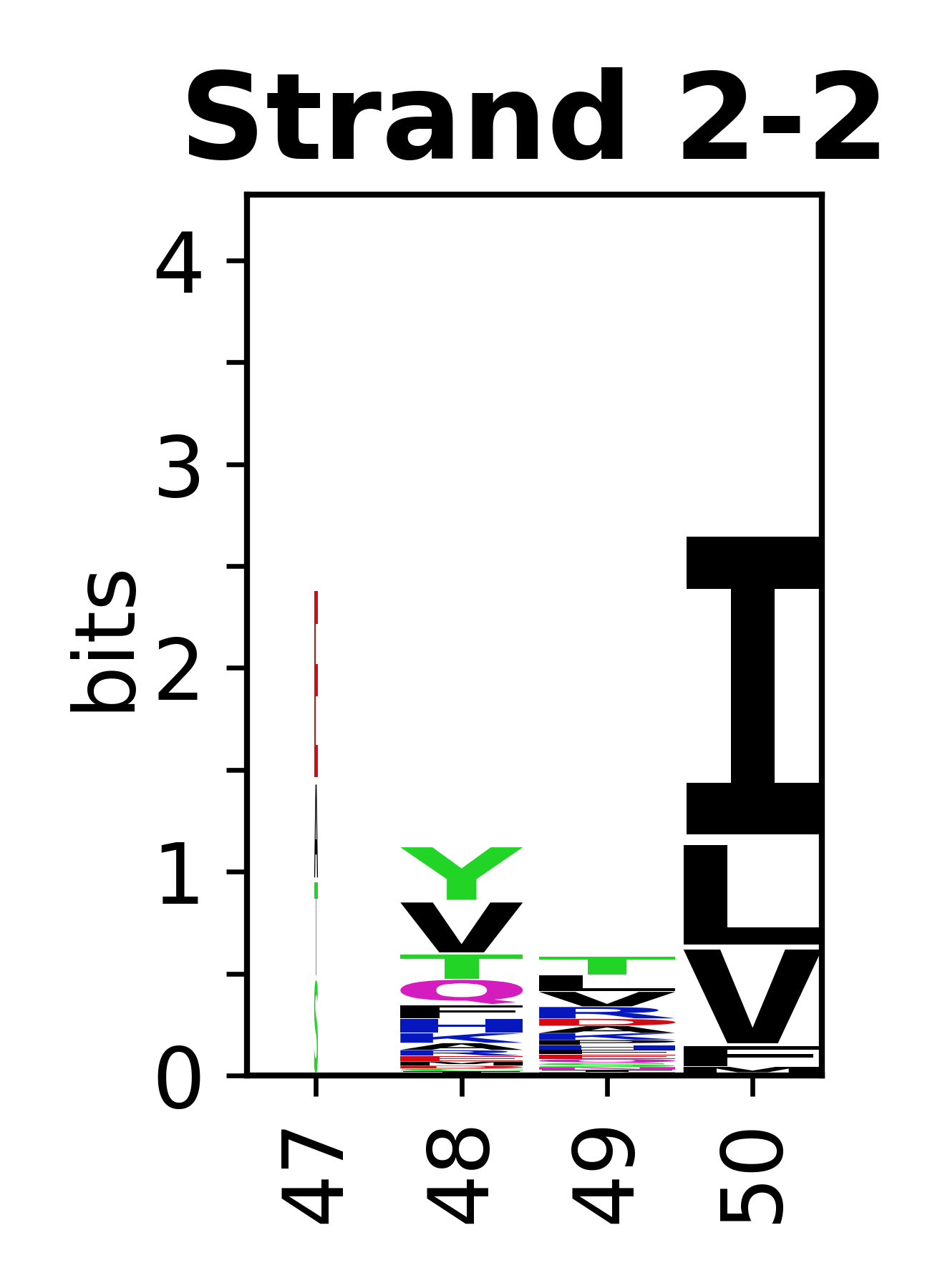
The amino acid sequences for each SSE can be aligned and used to produce a sequence logo. Where the sequence conservation is sufficient, we can establish a generic numbering scheme: the most conserved residue in helix X serves as its reference residue and is numbered as @X.50. The remaining residues in the helix are numbered accordingly. | |||
<gallery mode="packed" heights=140px caption="Helices"> | |||
File:SecStrAnnotator-cyp-sse-logo-A-.png | |||
File:SecStrAnnotator-cyp-sse-logo-A.png | |||
File:SecStrAnnotator-cyp-sse-logo-B.png | |||
File:SecStrAnnotator-cyp-sse-logo-B-.png | |||
File:SecStrAnnotator-cyp-sse-logo-B--.png | |||
File:SecStrAnnotator-cyp-sse-logo-C.png | |||
File:SecStrAnnotator-cyp-sse-logo-D.png | |||
File:SecStrAnnotator-cyp-sse-logo-E.png | |||
File:SecStrAnnotator-cyp-sse-logo-F.png | |||
File:SecStrAnnotator-cyp-sse-logo-F-.png | |||
File:SecStrAnnotator-cyp-sse-logo-G-.png | |||
File:SecStrAnnotator-cyp-sse-logo-G.png | |||
File:SecStrAnnotator-cyp-sse-logo-H.png | |||
File:SecStrAnnotator-cyp-sse-logo-I.png | |||
File:SecStrAnnotator-cyp-sse-logo-J.png | |||
File:SecStrAnnotator-cyp-sse-logo-J-.png | |||
File:SecStrAnnotator-cyp-sse-logo-K.png | |||
File:SecStrAnnotator-cyp-sse-logo-K-.png | |||
File:SecStrAnnotator-cyp-sse-logo-K--.png | |||
File:SecStrAnnotator-cyp-sse-logo-L.png | |||
File:SecStrAnnotator-cyp-sse-logo-L-.png | |||
</gallery> | |||
<gallery mode="packed" heights=140px caption="Beta strands"> | |||
File:SecStrAnnotator-cyp-sse-logo-1-0.png | |||
File:SecStrAnnotator-cyp-sse-logo-1-1.png | |||
File:SecStrAnnotator-cyp-sse-logo-1-2.png | |||
File:SecStrAnnotator-cyp-sse-logo-1-3.png | |||
File:SecStrAnnotator-cyp-sse-logo-1-4.png | |||
File:SecStrAnnotator-cyp-sse-logo-1-5.png | |||
File:SecStrAnnotator-cyp-sse-logo-2-1.png | |||
File:SecStrAnnotator-cyp-sse-logo-2-2.png | |||
File:SecStrAnnotator-cyp-sse-logo-3-1.png | |||
File:SecStrAnnotator-cyp-sse-logo-3-2.png | |||
File:SecStrAnnotator-cyp-sse-logo-3-3.png | |||
File:SecStrAnnotator-cyp-sse-logo-4-1.png | |||
File:SecStrAnnotator-cyp-sse-logo-4-2.png | |||
File:SecStrAnnotator-cyp-sse-logo-5-1.png | |||
File:SecStrAnnotator-cyp-sse-logo-5-2.png | |||
File:SecStrAnnotator-cyp-sse-logo-6-1.png | |||
File:SecStrAnnotator-cyp-sse-logo-6-2.png | |||
</gallery> | |||
<br style="clear:both" /> | <br style="clear:both" /> | ||
Latest revision as of 09:43, 16 July 2020
SecStrAnnotator Suite provides scripts (Python, R) for batch annotation of the whole family and analysis of the annotation results.
Procedure
[edit]Data preparation
[edit]The directory scripts/secstrapi_data_preparation/ contains a pipeline for annotating the whole protein family, including:
- downloading the list of family members defined by CATH and Pfam,
- downloading their structures,
- selecting a non-redundant set,
- annotation,
- multiple sequence alignment for individual SSEs,
- formatting into SecStrAPI format,
- formatting into TSV format for further analyses.
The whole pipeline can be executed by scripts/SecStrAPI_pipeline.py
Example usage:
python3 scripts/SecStrAPI_pipeline.py scripts/SecStrAPI_pipeline_settings.json --resume
Before running, modify the settings in SecStrAPI_pipeline_settings.json to set your family of interest, annotation template, data directory etc (see README.txt for more details).
Data analysis
[edit]The directory scripts/R_sec_str_anatomy_analysis/ contains a pipeline for statistical analysis of the annotation results on the whole protein family, including:
- reading the annotation results from TSV,
- generating plots,
- performing statistical test to compare eukaryotic and bacterial structures (or any two sets of structures).
Example usage:
- Launch
rstudiofrom the said directory - In
sec_str_anatomy.R, set DATADIR to the path to your annotation data created in #Data preparation - In
sec_str_anatomy_settings.R, modify the family-specific settings (list of helices and strands) - Run
sec_str_anatomy.Rline by line
Example case study: Cytochromes P450
[edit]Data
[edit]For the Cytochrome P450 family, structures of 1855 protein domains are available, located in 1012 PDB entries (updated on 7 July 2020). The analysis was performed on a non-redundant subset containing 183 protein domains.
The data are available here (structural files not included because of their size).
Occurrence of SSEs
[edit]The occurrence describes in what percentage of the structures a particular SSE is present.
Length of SSEs
[edit]The length of an SSE is measured as the number of residues. The following violin plots show the distribution of length for each SSE.
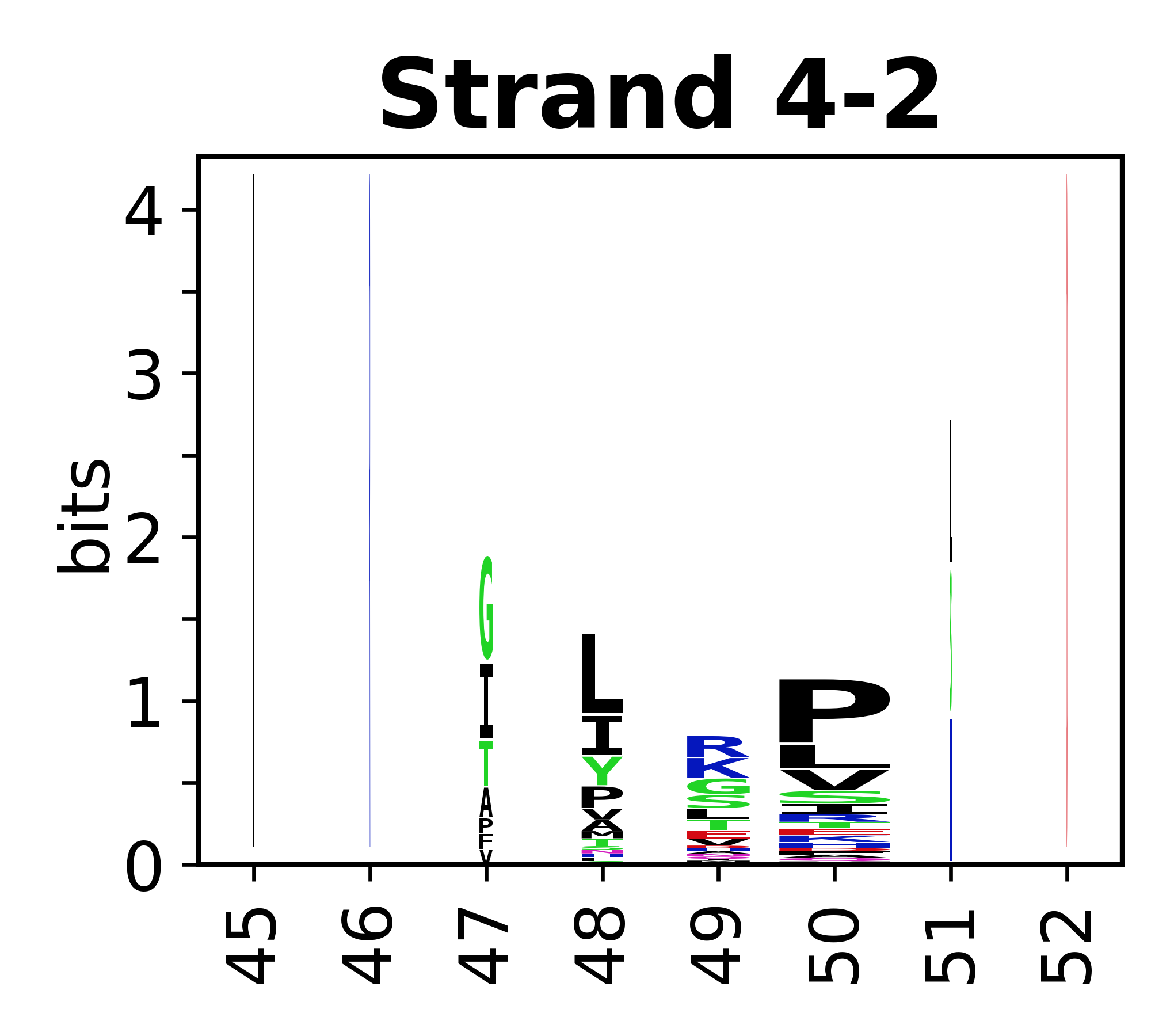
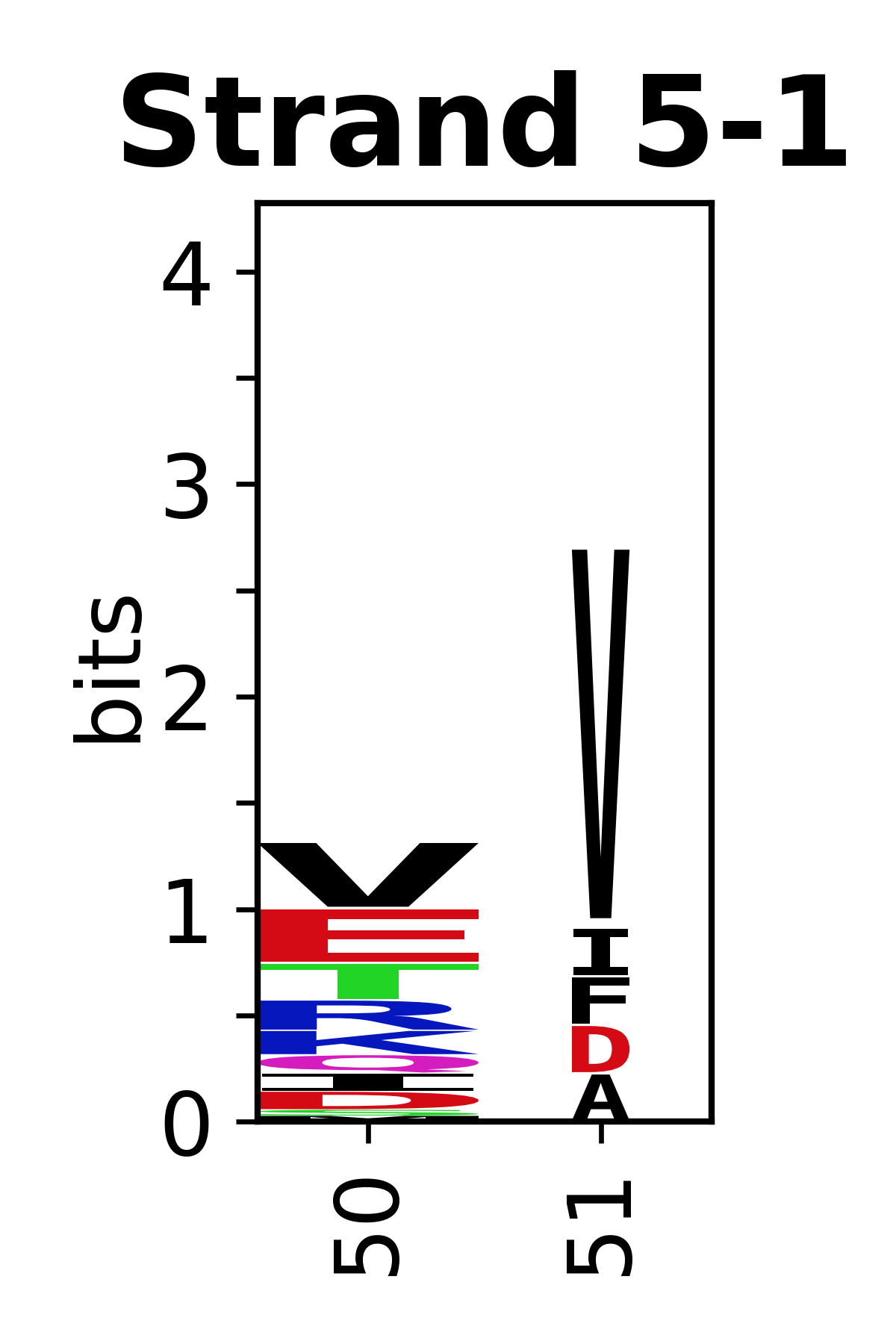
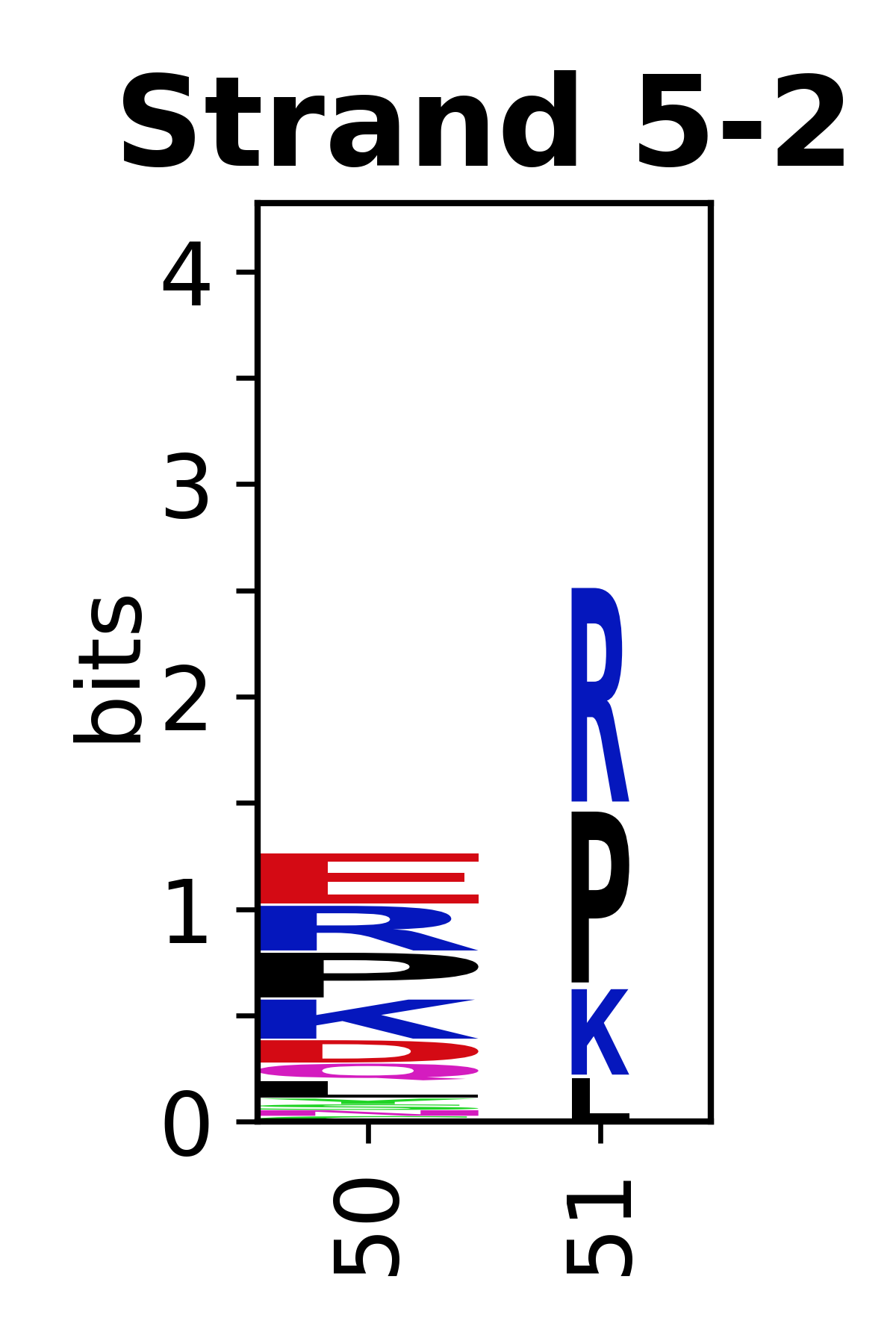
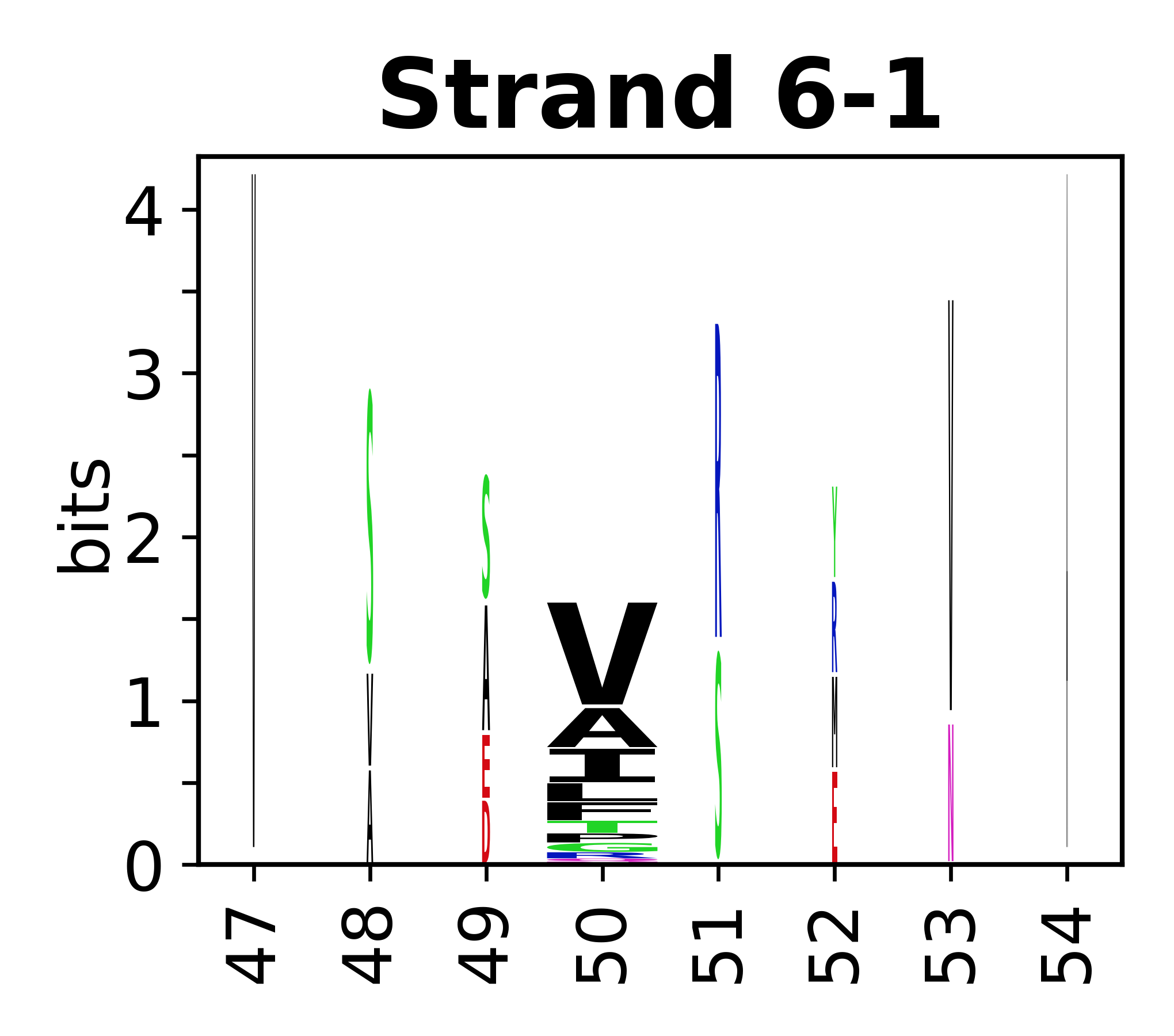
Sequence of SSEs
[edit]The amino acid sequences for each SSE can be aligned and used to produce a sequence logo. Where the sequence conservation is sufficient, we can establish a generic numbering scheme: the most conserved residue in helix X serves as its reference residue and is numbered as @X.50. The remaining residues in the helix are numbered accordingly.
- Helices
- Beta strands