ValidatorDB
The advancement of research in structural biology has provided a large body of structural data deposited in various databases. One great example is the Protein Data Bank (PDB), which has been growing exponentially, and which currently consists of more than 100,000 structures of biomolecules and their complexes. Such large bodies of data, especially accumulated over a short period of time using high throughput techniques, will inherently be plagued by various problems.
Validation arose as a major issue in the structural biology community when it became apparent that some published structures contained serious errors, and only sometimes it is properly explained(e.g., due to insufficient electron density in a certain area). Structural databases generally require that the new submissions be checked prior to acceptance. The tools employed for presubmission validations work fairly well for residues like amino acids or nucleotides. However, an essential step in the validation process is checking the ligand structure, because ligands play a key role in protein function, and also because they are the main source of errors in structures. Ligand validation, as well as the validation of uncommon residues, are very challenging tasks, because of the high diversity and nontriviality of their structure, and the general lack of information about correct structures. Therefore, software tools focused on ligand validation were developed relatively recently, <ref name="Lutteke_2004"/><ref name="Kleywegt_2007"/>, and the topic is still under active development<ref name="ftp"/>. These tools are able to validate one or more structures (even thousands of structures), but they are not able to provide the broad scientific community with a more complex image of the quality of structures in dedicated and well established structural databases. For example, a general overview and corresponding statistical evaluation of validation results for residues and ligands in the entire PDB is not yet available, despite the exponential growth of the PDB and the development of structural validation tools in recent years.
We had recently developed MotiveValidator<ref name="Varekova_2014"/>, an interactive platform for the speedy validation of ligands, residues and fragments using a novel, straightforward approach based on the validation of residue annotation. MotiveValidator employs advanced algorithms for the detection and comparison of structural motifs<ref name="Sehnal_2012"/>, along with tools for chirality verification<ref name="Boyle_2011" /> and interactive visualization of 3D structures<ref name="web"/>. Using MotiveValidator, we further created ValidatorDB, a comprehensive resource of validation results for residues and ligands in the Protein Data Bank. Along with validation results for individual residues and ligands, ValidatorDB also provides a summary and statistical evaluation of the validation results at various levels of detail within the PDB. Thus, ValidatorDB offers a comprehensive overview of the quality of the ligand structures in the entire PDB. ValidatorDB contains precomputed validation results for ligands and residues in the Protein Data Bank. The database is updated on a weekly basis.
The residues deemed relevant for validation are all ligands and residues with reasonable size (more than six heavy atoms), with the exception of standard amino acids and nucleotides. The validation is performed using MotiveValidator, and the residue models from wwPDB Chemical Component Dictionary (wwPDB CCD) are used as reference structures for validation.
Availability and technical details
Where to find ValidatorDB
ValidatorDB is freely available online since May 2014 at http://ncbr.muni.cz/ValidatorDB. There is no login requirement for accessing ValidatorDB.
What you need in order to run MotiveValidator
- Up to date internet browser with WebGL support.
- JavaScript enabled.
Check it out, if your browser is WebGL and Javascript compliant.
How to get around the web page
For the quick tour on using ValidatorDB service, please click the Guide button in the upper right corner and follow instructions.
Before moving on to more extensive descriptions of features, it is important to clearly establish the meaning of a few key terms and principles within the ValidatorDB environment. See Terminology.
Basic Principles
Residues and ligands relevant for validation
As mentioned in section 1, well studied residues like amino acids and nucleotides are routinely validated upon submission of new structures to the PDB. Furthermore, reports of the quality of their structure are already accessible. The challenge addressed by ValidatorDB lies in providing access to validation results for residues other than the well studied amino acids and nucleotides. This generally includes ligands and uncommon residues (e.g. substituted amino acids), which exhibit high diversity and nontriviality in their structure, and for which there is generally much less information regarding correct structures. Thus, within the ValidatorDB environment, we further refine the meaning of the terms residue and ligand to refer to residues and ligands relevant for validation. Specifically, these are all ligands and residues with reasonable size (more than six heavy atoms), with the exception of amino acids and nucleotides. All other features of the terms residue and ligand described in Terminology are maintained. Henceforth, all references to residues and ligands in the ValidatorDB web pages (including Wiki and tutorial) have the meaning of residues and ligands relevant for validation. The PDB currently holds over 170,00 residues and ligands relevant for validation.
Validation
Template:Columns-start Column 1 here Template:Column Column 2 here Template:Column Column 3 here Template:Columns-end
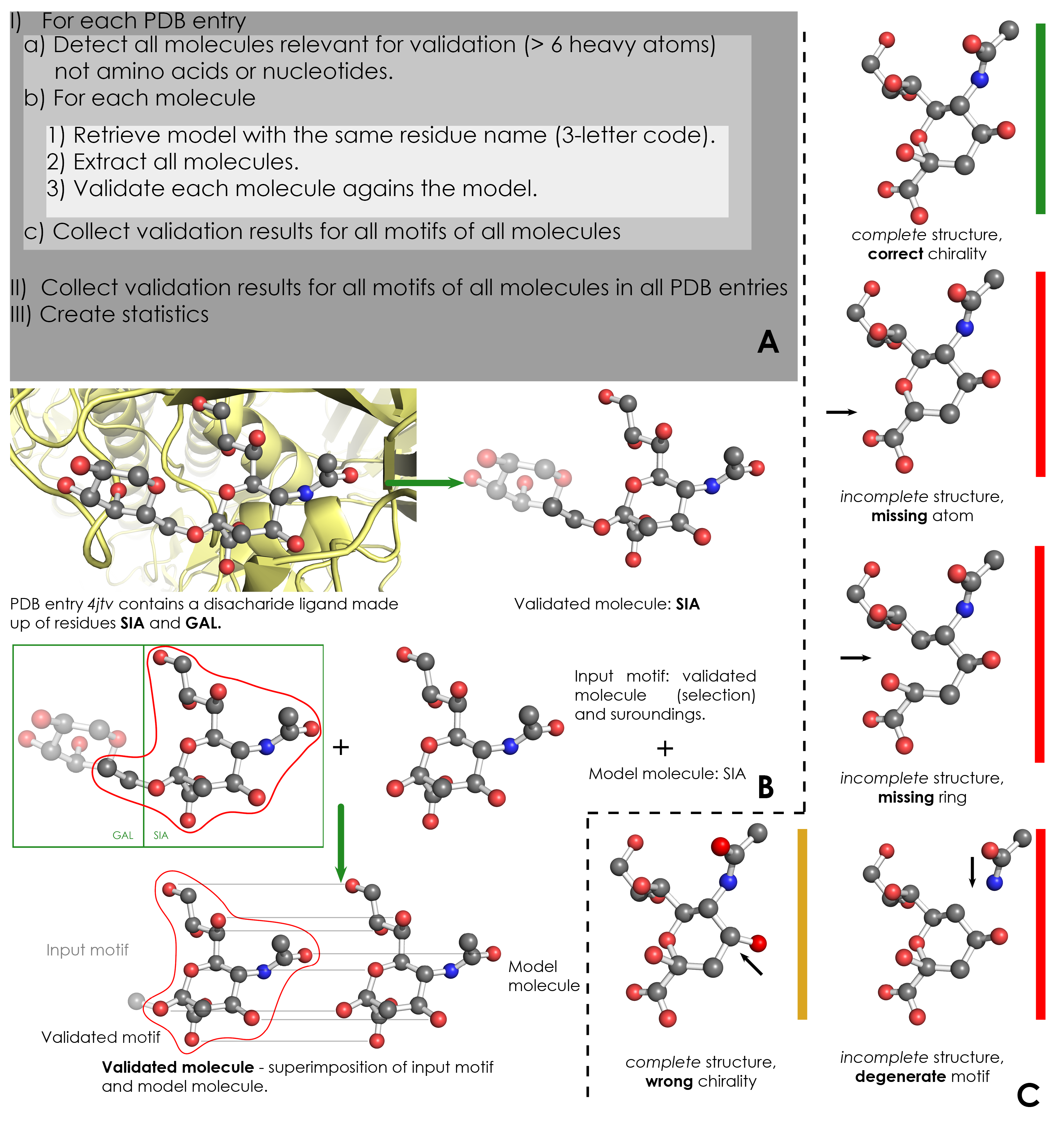
As stated in section 1, the validation results stored in ValidatorDB are updated every week. Within the ValidatorDB environment, the term validation refers to the process of determining whether a residue or ligand is structurally complete and correctly annotated. This means checking if the topology and chirality of each motif of a validated residue (Terminology) correspond to those of the model residue (Terminology) with the same name as the validated residue. The validation of residues and ligands in the entire PDB takes place in a few distinct steps. First, for each PDB entry, the residues which are relevant for validation are detected based on their name (3-letter code) and number of atoms (more than 6 heavy atoms). Amino acid residues and nucleotides are excluded based on their residue name. Then, for each validated residue, the corresponding model (same 3-letter code as the validated residue) is retrieved from wwPDB CCD, and each motif of this residue is validated against the model.
ValidatorDB is then built as the collection of validation results for all motifs of all residues in all PDB entries. The validation of each motif against the model residue can be illustrated on a galactose (GAL) motif from the PDB entry 1bzw. The validated residue GAL is extracted form PDB entry 1bzw in the form of an input motif , which contains all the atoms of the validated residue, together with all atoms found within one or two bonds of any atom from the validated residue(surroundings). Then, by superimposing the input motif and model residue, the validated motif is obtained as the subset of atoms in the input motif which have a correspondent atom in the model residue. Comparing each atom and bond in the validated motif to those in the model residue produces the validation results.
Validation results
Incomplete Structures
For each validated motif, ValidatorDB contains several types of results. Since the evaluation of the validated motif relies on comparing all atoms and bonds in the validated motif to those in the model residue, the first results that can be encountered are errors. Namely:
- Missing atoms: an atom in the model residue has no corresponding atom in the validated motif.
- Missing rings: at least one missing atom originates from cycles (rings).
- Wrong chirality: an atom from the validated motif has different chirality than the corresponding atom from the model residue.
- Wrong chirality (planar): the chirality error was found on a planar chiral center. Because of their spacial distribution, planar chiral centers are very sensitive even to small perturbations in the position of the substituents. Therefore, some of the errors reported here might not be significant.
- Uncertain chirality: the presence of unusual bonds may cause an improper evaluation of chirality.
Complete Structures
Chirality is only evaluated for those motifs which are complete. This is because the absence of some atoms can prevent the proper evaluation of chirality on the chiral centers present in the validated motif. Therefore, note that all motifs which are counted in the Wrong chirality category are in fact complete. At the same time, the motifs with no missing atoms and no chirality error are actually counted in a category called Correct chirality.
Warnings
Suspicious discrepancies between the atoms and inter-atomic bonds in the validated motif and in the model residue are reported as warnings. Namely:
- Substitution: an atom from the validated motif is of a different chemical element than the corresponding atom in the model residue (e.g. O mapped to N). This happens often at linkage sites.
- Different naming: an atom from the validated motif has a different PDB atom name than the corresponding atom from the model residue (e.g. the C1 atom mapped to the C7 atom). This happens often when the original PDB files were produced by different software.
- Foreign atom: an atom from the model residue was mapped to an atom from outside the validated residue (i.e. from its surroundings).
- Alternate locations: in the original PDB file, the validated residue contains atoms which were given in alternate locations (i.e., most probably different rotamers). Only the first rotamer was considered during validation.
- Zero model RMSD: the superimposition between the model residue and the validated motif has a root mean square deviation of zero, i.e., the validated motif is identical to the model residue used as reference.
Disabling discrepancies between the atoms and inter-atomic bonds in the validated motif and in the model residue are reported as processing errors, and such motifs are not validated.
As a general rule, in the ValidatorDB interface, errors are marked in red (missing atoms) or dark yellow (wrong chirality), correct structures in green, and warnings in cyan.
Database contents
ValidatorDB contains precomputed validation results for ligands and residues in the Protein Data Bank. The database is updated on a weekly basis. The validation is performed using Validator, and the residue models from wwPDB CCD are used as reference templates for validation. All residues of significant size (a minimum of 6 heavy atoms) have been included in ValidatorDB, with the exception of amino acids and nucleotides, which are checked thoroughly upon submission of the structure to the PDB, and thus do not require additional validation.
The validation results available in ValidatorDB inform whether each motif (occurrence, instance) of a ligand or residue in the PDB exhibits the appropriate topology and stereochemistry expected from its annotation (3-letter code), or how it differs from this annotation. Additionally, all issues related to incorrect or suspicious topology and stereochemistry are explicitly described in a comprehensive and intuitive manner (e.g., location of missing atoms or chirality inversions).
ValidatorDB is organized on two main levels, namely PDB-wide results (synopsis page), and results restricted to specific residues of interest (specifics page). The two levels present the same type of validation results (as described in section 2.7), although the available features differ to some extent (e.g., the specifics page allows 3D visualization of motifs). We shall describe each level of the database in detail below.
Synopsis page
The ValidatorDB synopsis page(Figures1A, 3) contains a brief description of ValidatorDB, along with information about the last database update (date and number of structures that have been processed during the validation). Specifically, in May 2014, over 100,000 PDB entries had been processed, containing over 230,000 motifs of 17,000 residues relevant for validation. Additionally, the synopsis page allows to access the validation results for specific residues of interest via the LookUp bar (Figure 1A). Simply type a comma separated list of residue names (3-letter codes) into the LookUp bar, and you will be redirected to the specifics page containing validation results for the residues you requested. If you specify a list of PDB IDs (4-letter codes)instead, then the corresponding specifics page will contain validation results for all relevant residues and ligands in the PDB entries you specified. See section 3.2 for a description of the contents of the specifics page, and how to interpret these contents. The ValidatorDB synopsis page further provides access to various data sets of PDB-wide validations via 3 different tabs, namely Overview, Details by Residue, and Details by PDB entry. A full description of each of these tab is given below (sections 2.1.1-2.1.3).
Overview
The Overview tab of the synopsis page provides a very general statistical evaluation of results across the entire PDB in graphical form (Figures 1A,3A). The elements of the graph represent percentages of the total number of motifs (over 200,000) of residues relevant for validation. A graphic element will be displayed in the Overview graph only if it represents at least 0.5% of the total number of motifs. Each element of the graph is described in a tool tip, but note that here the term residue actually refers to occurrence of residue(motif).
The elements of the graph can be assigned to roughly 6 categories, depending on which kind of information they contain (e.g., incomplete residue, chirality issues, warnings, etc). The categories are marked by different colors. Most of the graph elements have been explained in section 3.7 of this manual. The additional elements are Analyzed, which refers to the total number of motifs that could be processed, Missing Atoms or Rings, which is the sum of Missing (Only) Atoms and Missing Rings, and Has All Atoms and Rings, which is the total number of complete residues.
Details by Residue
The Details by Residue tab (Figure 3B) contains an interactive table summarizing the results for each residue validated across the entire PDB. Each row corresponds to one residue, identified by its residue name (3-letter code). The information in the table is organized according to the validation results as presented in section 2.3 of this manual. The color coding for the table header and the font inside the table is the same as in the categories defined in the Overview tab. Each element of the table header is described in a tool tip, but note that here the term residue actually refers to occurrence of residue (motif).
The table is interactive. Clicking on any element in the table header allows to sort the table entries according to that element. Click on any residue name in order to access the ValidatorDB specifics page with detailed validation results for that residue (see section 3.2).
The filter at the top right corner allows to retrieve the table row with a specific residue. Simply type the residue name into the filter. All results can be downloaded in .csv format using the download button at the top left corner.
Details by PDB entry
The Details by PDB Entry tab contains an interactive table summarizing the results for all residues validated in each PDB entry. Each row corresponds to one PDB entry, identified by its PDB ID (4-letter code). The information in the table is organized according to the validation results as presented in section 2.3 of this manual. The color coding for the table header and the font inside the table is the same as in the categories defined in the Overview tab. Each element of the table header is described in a tool tip, but note that here the term residue actually refers to occurrence of residue (motif).
The table is interactive. Clicking on any element in the table header allows to sort the table entries according to that element. Click on any PDB in order to access the ValidatorDB specifics page with detailed validation results for all residues in that PDB entry (see section 3.2).
The filter at the top right corner allows to retrieve the table rows with a specific residue, or the table rows with selected PDB IDs. Simply type the residue name or PDB ID into the filter. All results can be downloaded in .csv format using the download button at the top left corner.
References
<references> <ref name="Lutteke_2004">Lütteke,T. and von der Lieth,C.-W. (2004) pdb-care (PDB carbohydrate residue check): a program to support annotation of complex carbohydrate structures in PDB files. BMC Bioinformatics, 5, 69.</ref>
<ref name="Kleywegt_2007">Kleywegt,G.J. and Harris,M.R. (2007) ValLigURL: a server for ligand-structure comparison and validation. Acta Crystallogr. D. Biol. Crystallogr., 63, 935–8.</ref>
<ref name="ftp">ftp://ftp.ebi.ac.uk/pub/databases/pdb/validation_reports/</ref>
<ref name="Varekova_2014">Vařeková,R.S., Jaiswal,D., Sehnal,D., Ionescu,C.-M., Geidl,S., Pravda,L., Horský,V., Wimmerová,M. and Koča,J. (2014) MotiveValidator: interactive web-based validation of ligand and residue structure in biomolecular complexes. Nucleic Acids Res., 12, 227–233.</ref>
<ref name="Sehnal_2012">Sehnal,D., Vařeková,R.S., Huber,H.J., Geidl,S., Ionescu,C.-M., Wimmerová,M. and Koča,J. (2012) SiteBinder: an improved approach for comparing multiple protein structural motifs. J. Chem. Inf. Model., 52, 343–59.</ref>
<ref name="Boyle_2011">O’Boyle,N.M., Banck,M., James,C. a, Morley,C., Vandermeersch,T. and Hutchison,G.R. (2011) Open Babel: An open chemical toolbox. J. Cheminform., 3, 33.</ref> <ref name="web">http://www.chemdoodle.com</ref> </references>