PatternQuery:Service Organization
Appearance
Motive Query service is aimed for querying large datasets of structures from the Protein Data Bank. For the perfecting your query or querying smaller datasets or even user-uploaded data, please use MotiveQuery Explorer instead.
If you would like to run MotiveQuery at your home institution, please download a command-line version. The following visual will guide you through the process of submitting you calculation so as the interpreting results.
Synopsis Page
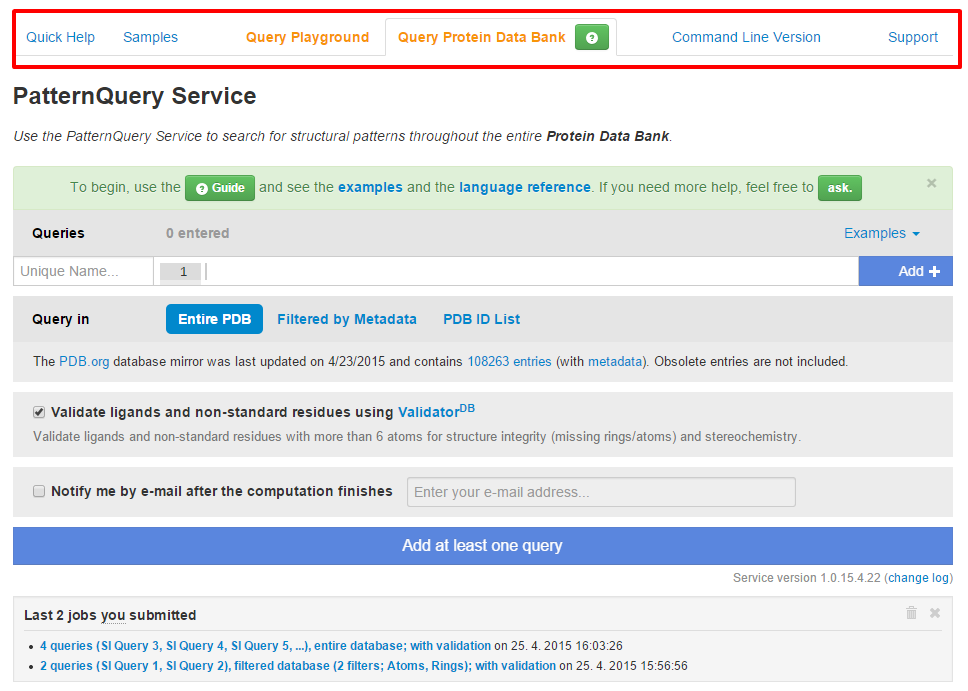
The MotiveQuery synopsis page is divided into a number of tabs:
- Quick Help and Samples tab provides information to help you orient better.
- Support provides a simple user interface for asking questions related to a MotiveQuery language or service. We do our best to come back to your questions as soon as possible. Query Playground represents an interface for the MotiveQuery Explorer application where you can tune up your queries before doing the database-wide searches.
- Query Protein Data Bank is a simple interface for creating a database wide MotiveQuery calculation.
- Command Line Version enables you to download a MotiveQuery service and run it at your home institution over your in-house database in a supported format (*.pdb, *.cif, *.pqr, *.mol and *.sdf).
Job Submission
|
|
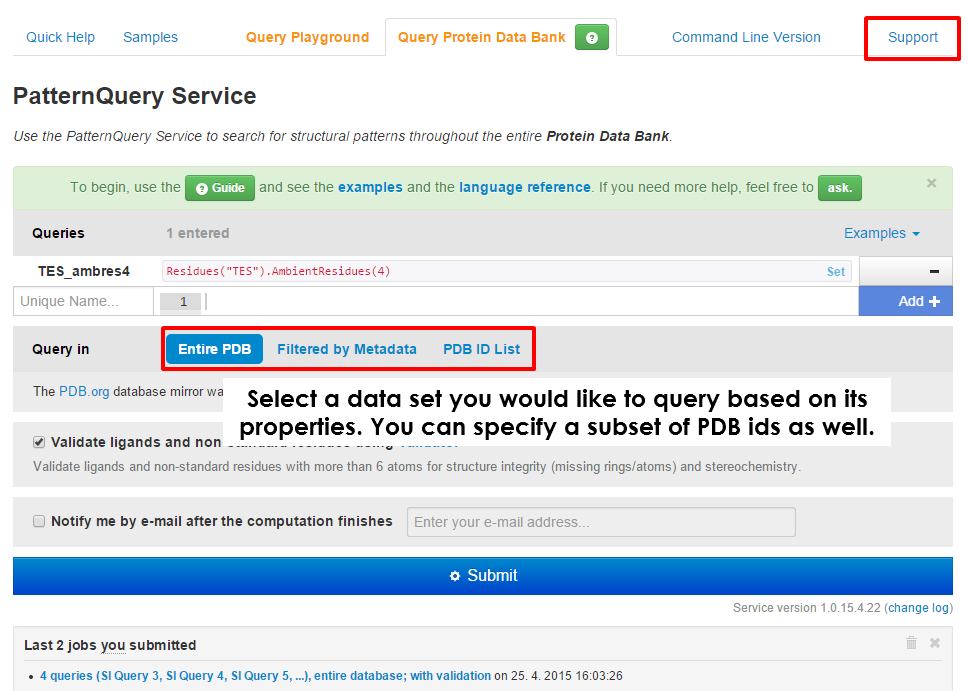
| The UI of the MotiveQuery Service job submission page allows user to submit a new query calculation. First time user can take a look at some of the example queries. If you have run any calculations with MotiveQuery already, you can browse the results. | At first, decide whether you would like to query an entire Protein Data Bank, its subset based on a variety of different properties, or you have a list of PDB entries of your interest. |
|
|
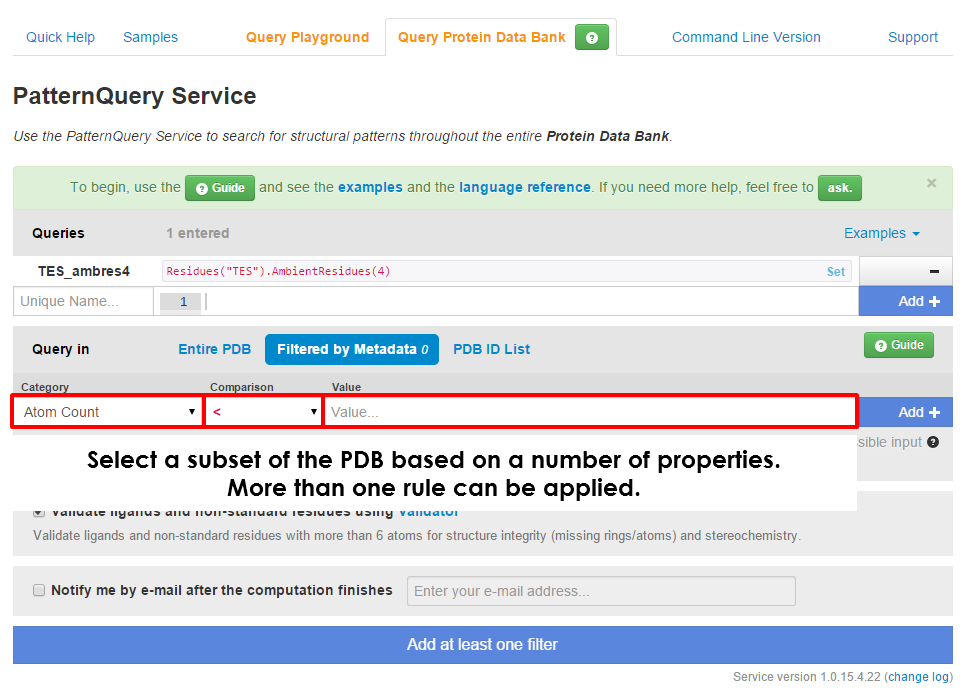
| You can to query a subset of Protein Data Bank based on numerous metadata. At first select a property you would like to filter by, then select a comparison method and finally fill in the demanded value. Please read carefully hints and examples below each selected property. Different properties can have different means of providing input values. Note that you can also include a logical expressions. For example: S & !N in Atom property means that just structures containing a sulfur atom and none nitrogen atom will be considered. Multiple filters can be applied at the same time. | Once you select a data set, you can specify multiple queries which will be executed. If you are unsure on how to compose a query, try one of the provided examples. You can also take a look into our case studies or consult the language reference. In case none of these help, ask us a question via Support tab. We will be happy to answer you in the shortest possible time. Do not forget to click + Add after you finish composing query. |
| |
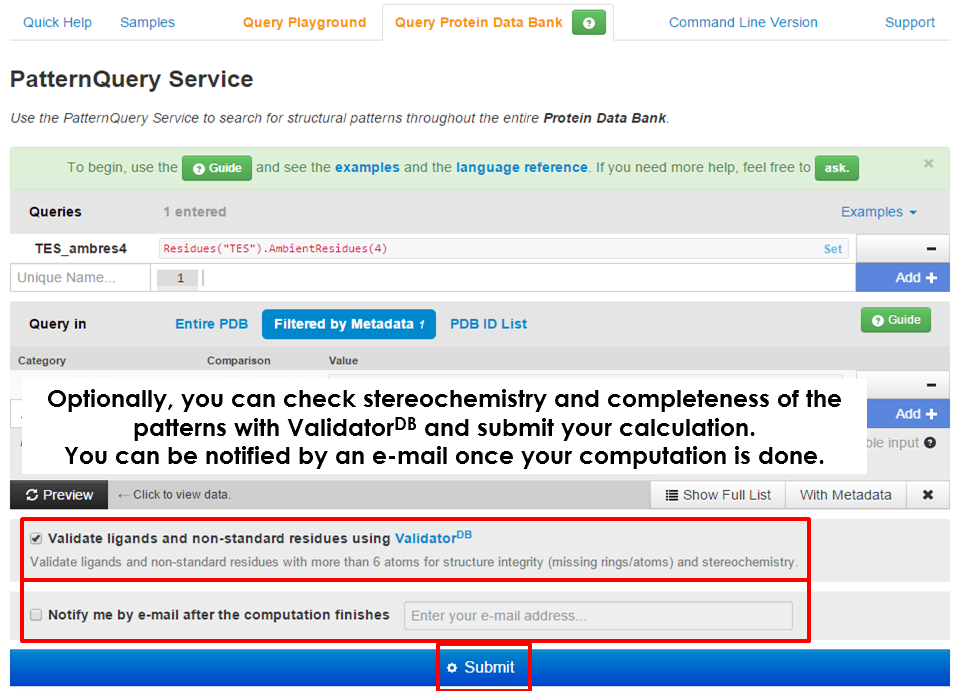
| As soon as you specify a query or queries and so data set, you can tick an option to validate all the ligands and non-standard residues for their structure integrity using ValidatorDB and, finally, submit the calculation. Based on the number of processed structures, the execution time can be in the range of minutes (thousands of structures) to an hour (whole Protein Data Bank). Bookmark the newly generated web address as the results will be available here upon the calculation finishes. |