ChargeCalculator:Interpretation of results: Difference between revisions
No edit summary |
No edit summary |
||
| Line 3: | Line 3: | ||
=Synopsis page= | =Synopsis page= | ||
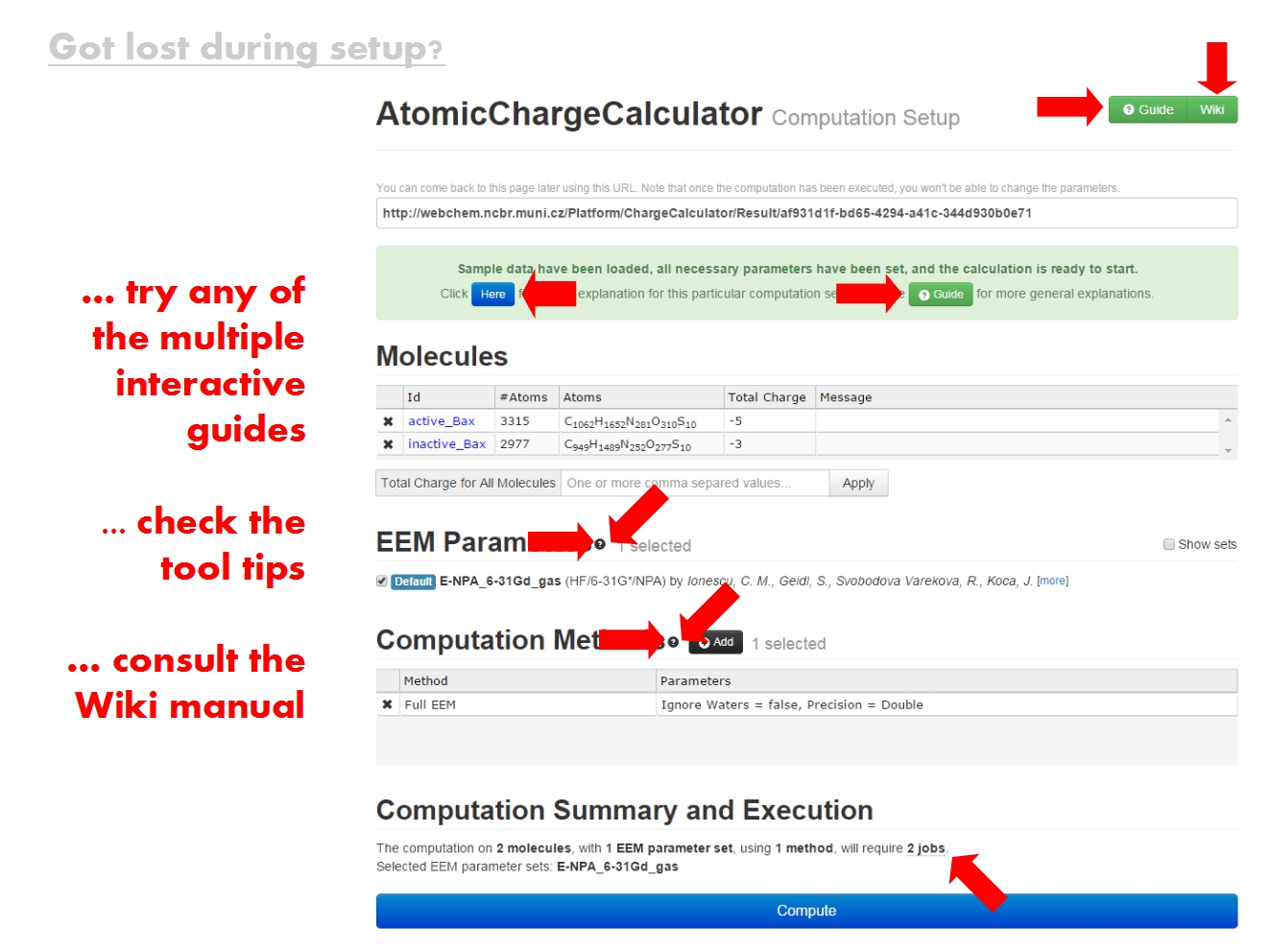
The '''ACC Synopsis page''' contains an overview of your computation, which might have consisted of multiple jobs for multiple molecules. | |||
Download all the results now (molecular structure files containing atomic charges, statistics of the results, information about all jobs), or return to this page later via this URL. You may also inspect the results directly in your web browser. | |||
Each <b>molecule</b> is identified according to the name of the input file, and comprises all structural elements in the input file, from simple compounds to biomacromolecular complexes made up of proteins, nucleic acids, ligands, ions, water, etc. For each molecule, your computation may have consisted of several jobs, in which case all the results will be available for comparison. Click on a molecule Id to visualize and analyze the results for this molecule in detail. | |||
{| class="wikitable" border="1" style="margin: 1em auto 1em auto;" | {| class="wikitable" border="1" style="margin: 1em auto 1em auto;" | ||
| Line 27: | Line 22: | ||
==Summary== | ==Summary== | ||
Overview of the calculation, including warnings. Basic information about the input molecule and the setup of the calculation. | |||
The ''molecule'' comprises all structural elements present in the <b>input file</b>, regardless of annotations. Molecular formula, describing the atomic composition. Total charge of the molecule, distributed across the constituent atoms based on the Electronegativity Equalization Method (EEM). Total duration of all jobs for this molecule. Individual job timing available for download with the rest of the results. | |||
[[File:ACC FirstTimeUserGuide 13.jpg|thumb|right|650px|Caption about Summary.]] | |||
Multiple ''jobs'' may have been run for this molecule. A job is defined based on the molecule and its total charge, along with the EEM parameter set and computation method used. Each job produces a distinct <b>set of charges</b> with a unique ID encoding the EEM parameter set, method and total molecular charge. If the original input file contained values of atomic charges, these are also stored in a charge set named according to the input file, and marked by <i>ref</i>. All charge sets are available for analysis in the remaining tabs. | |||
Warnings and errors generated while processing the input files and running the calculation. Each job will have its own list of errors and warnings. Jobs with errors generally do not produce any charge set, while jobs with warnings do. Warnings inform about some action taken under unexpected circumstances (e.g, defaulting to different parameters for certain atoms), or draw attention to aspects which may represent issues (e.g., insufficient number of H to satisfy all valences). | |||
<br style="clear:both" /> | <br style="clear:both" /> | ||
| Line 62: | Line 36: | ||
[[File:ACC_FirstTimeUserGuide 14.jpg|thumb|right|650px|]] | [[File:ACC_FirstTimeUserGuide 14.jpg|thumb|right|650px|]] | ||
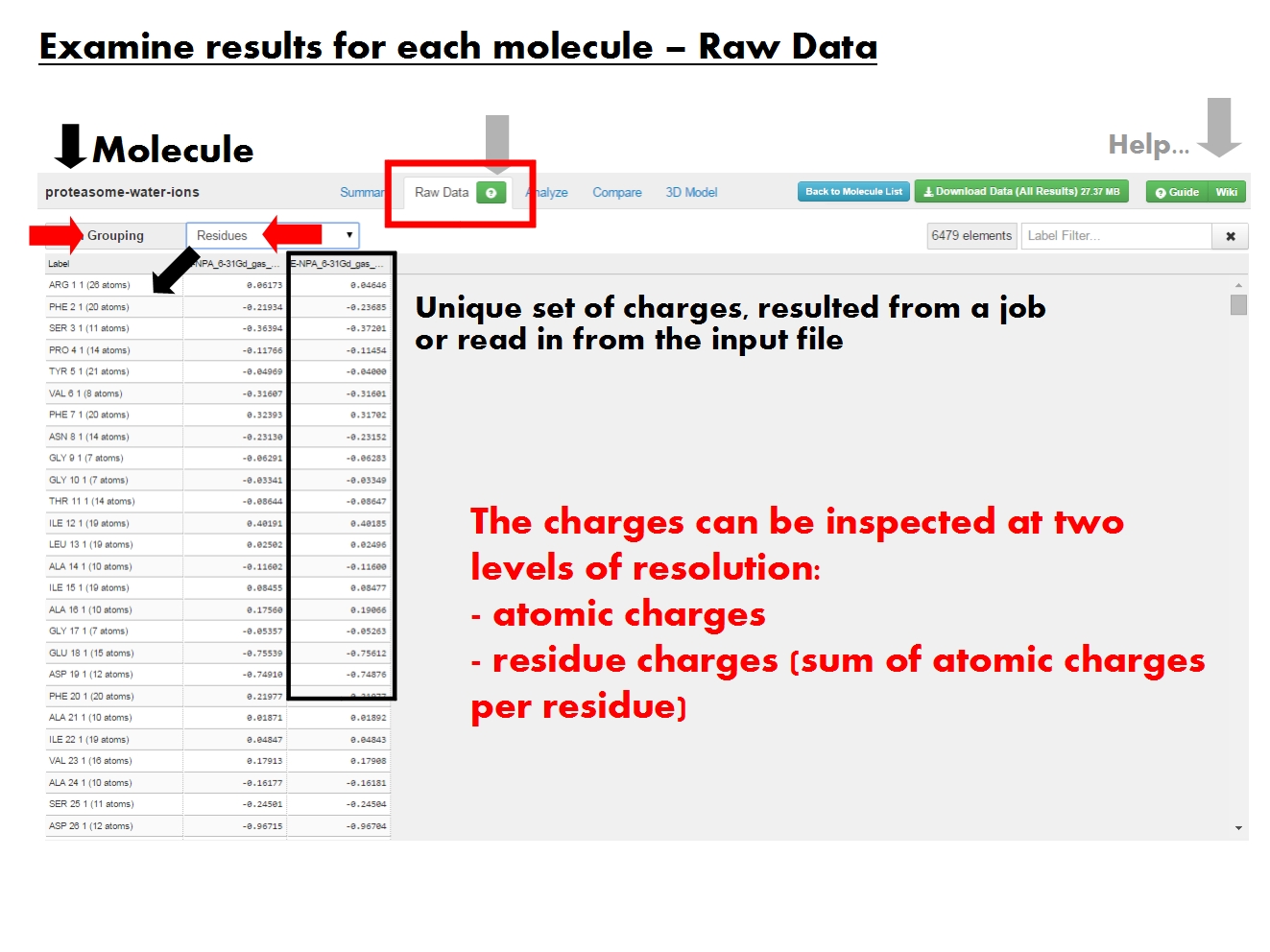
The ''raw data'' is available in tabular form, and refers to the actual values of atomic charges resulted from the EEM calculation, or read in from the input file. Residue charges may also be available. No additional statistics included here. Raw data available at more levels of resolution, termed ''atom grouping''. Pick ''Atoms'' to list atomic charges, or ''Residues'' to list residue charges (sum of atomic charges for each residue). Residue charges available only if residue annotations are present in the input file. | |||
Unique entry based on ''atom grouping''. Labels for ''Atoms'' contain the atom type, atom name, atom serial number and residue of origin. Labels for ''Residues'' contain the residue name, residue serial number, chain identifier and number of atoms in the residue. Entries can be sorted according to atom or residue serial number. | |||
Each job produced one set of charges, identified by the EEM parameter set, method and total molecular charge. Each column contains atom or residue charges resulted from a single job, or read from the input file (identified by the file name and marked by ''ref''. | |||
Filter table entries according to label. Regular expressions work. Counter records filter results. | |||
<br style="clear:both" /> | <br style="clear:both" /> | ||
| Line 85: | Line 49: | ||
[[File:ACC FirstTimeUserGuide 15.jpg|thumb|right|650px|]] | [[File:ACC FirstTimeUserGuide 15.jpg|thumb|right|650px|]] | ||
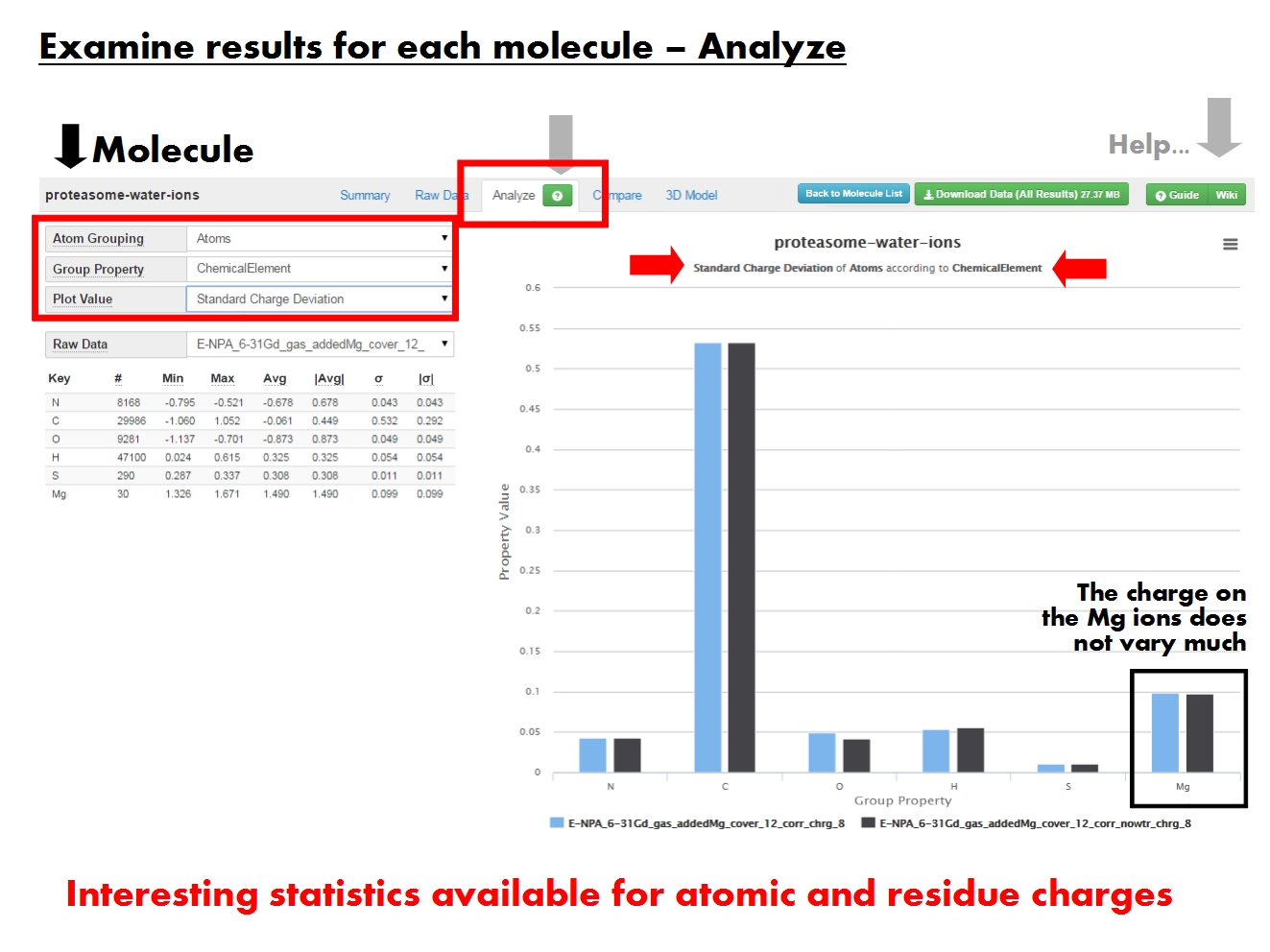
'''ACC''' provides several statistical descriptors for the results at more levels of resolution, in graphical and tabular form. | |||
Displays the ''value'' of a statistical descriptor for charges on ''groups of atoms'', according to a certain ''property''. For example, the ''minimum charge'' of ''atoms'' according to their ''chemical element''. Content of the graph dictated by the menus on the left. | |||
Each column represents the results from a single job (defined by EEM parameter set, computation method, and total molecular charge), or read in from the input file (identified by the file name and marked by ''ref''. | |||
Dictate contents of the graph to the right, and of the ''Raw Data'' table below. | |||
addStep("charges-aggregate-ag", //atom grouping and menu | addStep("charges-aggregate-ag", //atom grouping and menu | ||
Revision as of 08:51, 4 December 2014
bla bla bla
Synopsis page
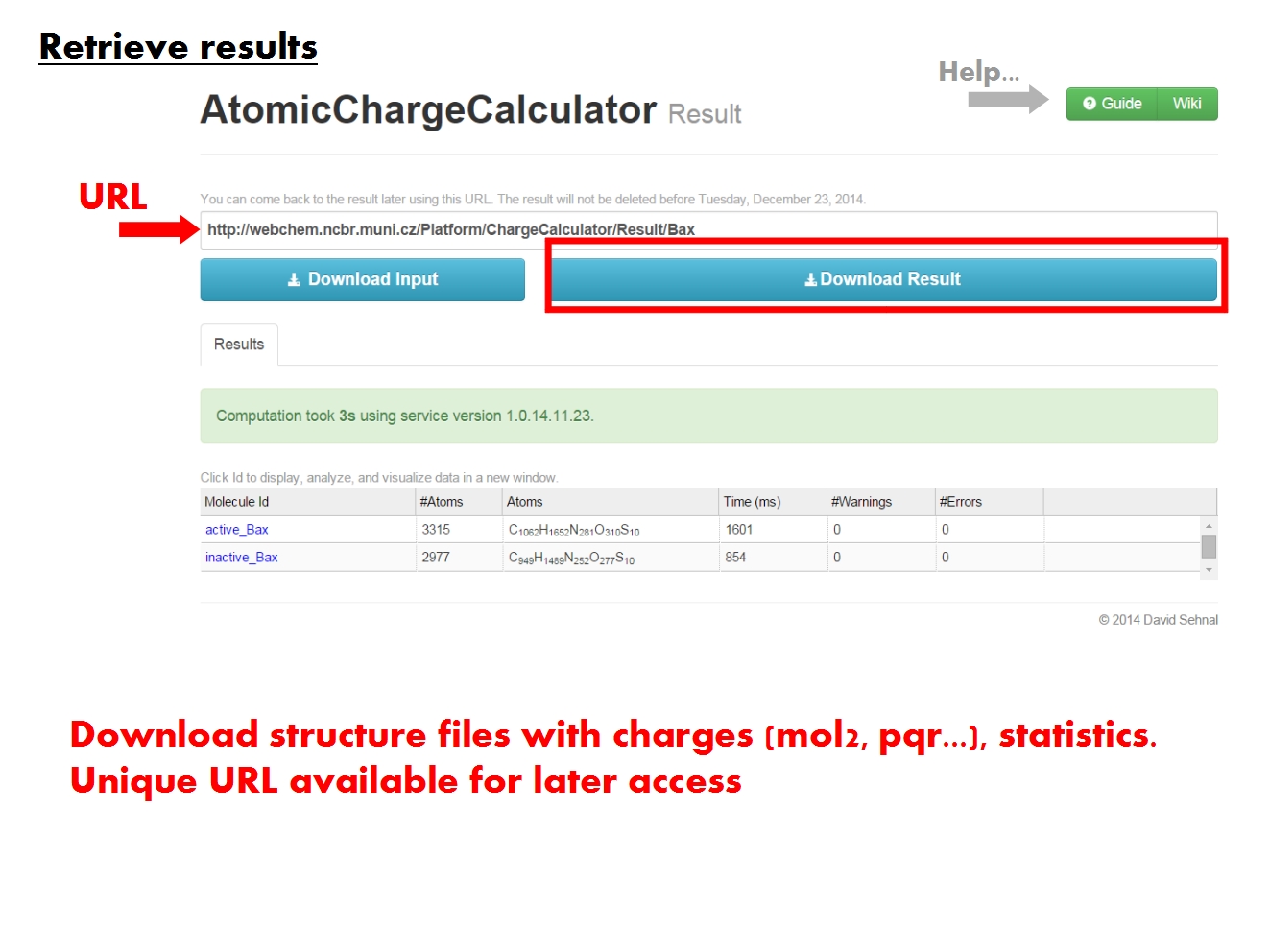
The ACC Synopsis page contains an overview of your computation, which might have consisted of multiple jobs for multiple molecules. Download all the results now (molecular structure files containing atomic charges, statistics of the results, information about all jobs), or return to this page later via this URL. You may also inspect the results directly in your web browser. Each molecule is identified according to the name of the input file, and comprises all structural elements in the input file, from simple compounds to biomacromolecular complexes made up of proteins, nucleic acids, ligands, ions, water, etc. For each molecule, your computation may have consisted of several jobs, in which case all the results will be available for comparison. Click on a molecule Id to visualize and analyze the results for this molecule in detail.
|
|
| Caption for download. | Caption for molecules. |
Specifics page
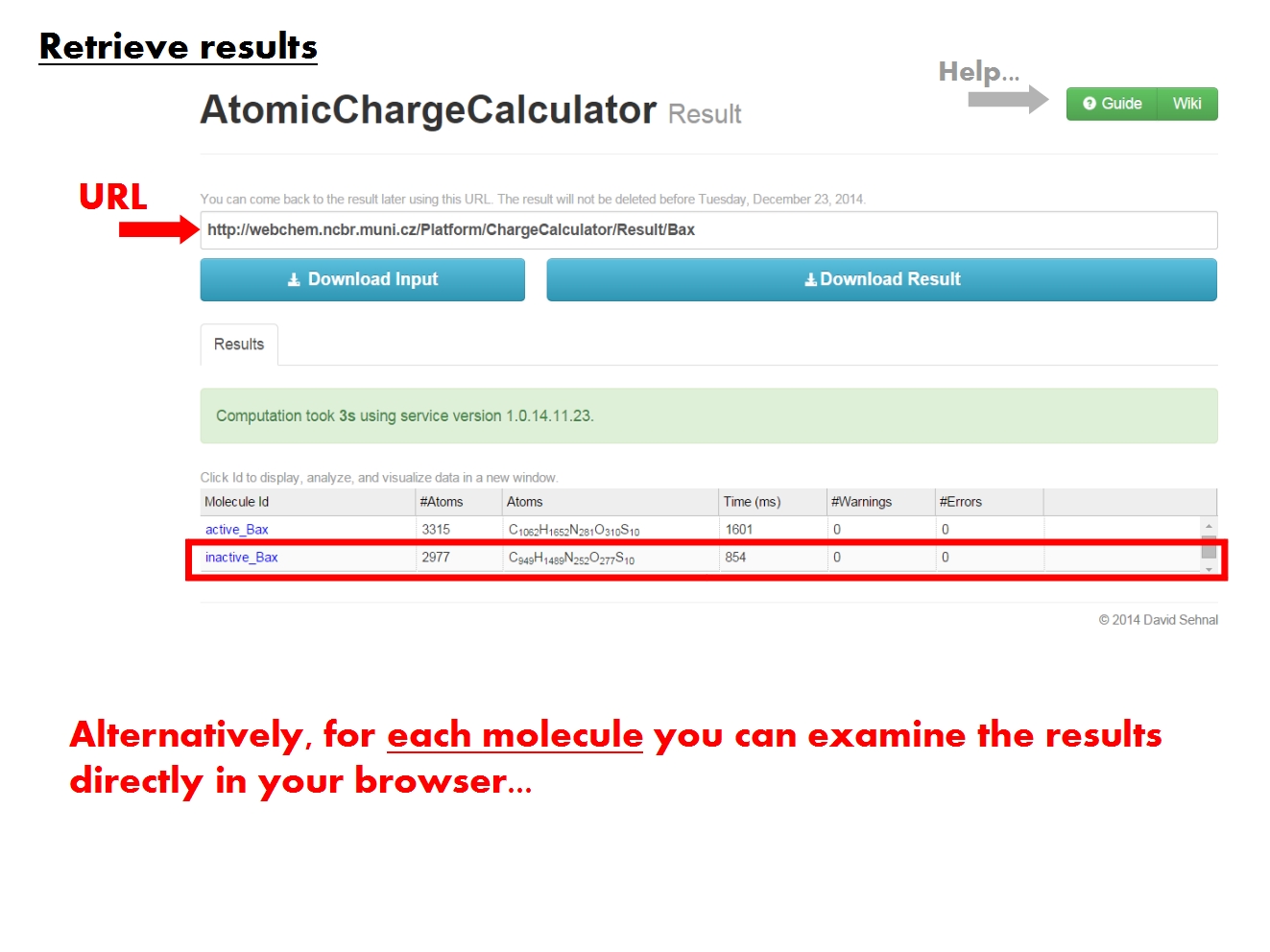
The ACC Specifics page contains the results of all atomic charge calculation jobs pertaining to a single molecule, identified according to the name of the original input file. Functional tabs allow to view and analyze various parts of the data directly in your browser.
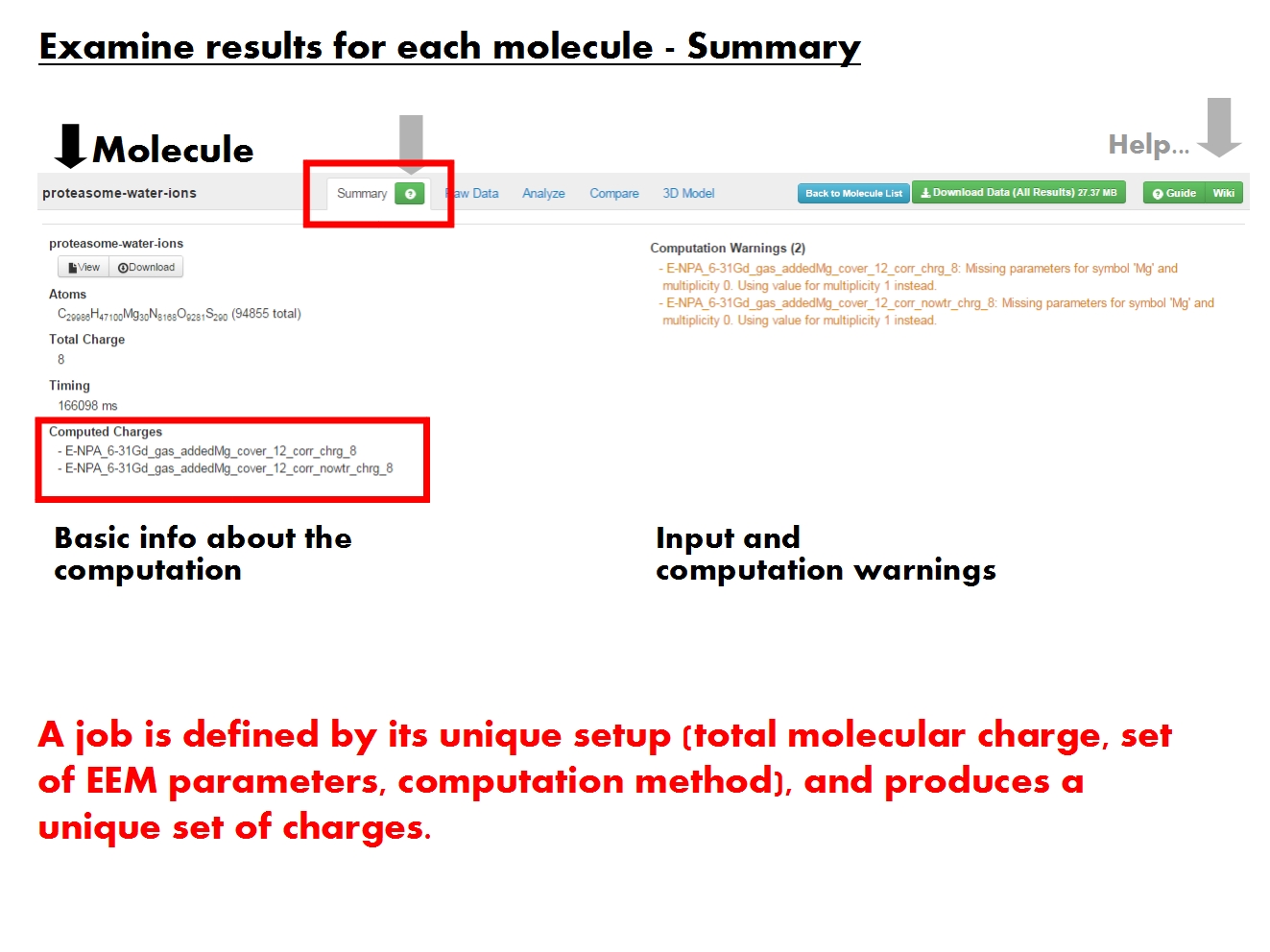
Summary
Overview of the calculation, including warnings. Basic information about the input molecule and the setup of the calculation. The molecule comprises all structural elements present in the input file, regardless of annotations. Molecular formula, describing the atomic composition. Total charge of the molecule, distributed across the constituent atoms based on the Electronegativity Equalization Method (EEM). Total duration of all jobs for this molecule. Individual job timing available for download with the rest of the results.
Multiple jobs may have been run for this molecule. A job is defined based on the molecule and its total charge, along with the EEM parameter set and computation method used. Each job produces a distinct set of charges with a unique ID encoding the EEM parameter set, method and total molecular charge. If the original input file contained values of atomic charges, these are also stored in a charge set named according to the input file, and marked by ref. All charge sets are available for analysis in the remaining tabs.
Warnings and errors generated while processing the input files and running the calculation. Each job will have its own list of errors and warnings. Jobs with errors generally do not produce any charge set, while jobs with warnings do. Warnings inform about some action taken under unexpected circumstances (e.g, defaulting to different parameters for certain atoms), or draw attention to aspects which may represent issues (e.g., insufficient number of H to satisfy all valences).
Raw Data
The raw data is available in tabular form, and refers to the actual values of atomic charges resulted from the EEM calculation, or read in from the input file. Residue charges may also be available. No additional statistics included here. Raw data available at more levels of resolution, termed atom grouping. Pick Atoms to list atomic charges, or Residues to list residue charges (sum of atomic charges for each residue). Residue charges available only if residue annotations are present in the input file.
Unique entry based on atom grouping. Labels for Atoms contain the atom type, atom name, atom serial number and residue of origin. Labels for Residues contain the residue name, residue serial number, chain identifier and number of atoms in the residue. Entries can be sorted according to atom or residue serial number.
Each job produced one set of charges, identified by the EEM parameter set, method and total molecular charge. Each column contains atom or residue charges resulted from a single job, or read from the input file (identified by the file name and marked by ref.
Filter table entries according to label. Regular expressions work. Counter records filter results.
Analyze
ACC provides several statistical descriptors for the results at more levels of resolution, in graphical and tabular form.
Displays the value of a statistical descriptor for charges on groups of atoms, according to a certain property. For example, the minimum charge of atoms according to their chemical element. Content of the graph dictated by the menus on the left.
Each column represents the results from a single job (defined by EEM parameter set, computation method, and total molecular charge), or read in from the input file (identified by the file name and marked by ref.
Dictate contents of the graph to the right, and of the Raw Data table below.
addStep("charges-aggregate-ag", //atom grouping and menu
"Atom Grouping gives the level of resolution. Pick Atoms to display atomic charges, or Residues to display residue charges (sum of atomic charges for each residue). Residue-based statistics available only if residue annotations present in input file.",
"right");
addStep("charges-aggregate-group", //group property and menu
"Group Property sets the specific property based on which the statistics are calculated for a certain Atom Grouping. For example, statistics on all atoms with a certain chemical element, or statistics on all polar residues.",
"right");
addStep("charges-aggregate-pv", //plot value and menu
"Plot Value refers to the specific statistical descriptor displayed in the graph. For example, the standard deviation of the charge on atoms with a certain chemical element.",
"right");
addStep("charges-aggregate-raw", //Raw data and menu and table
"Tabular representation of all statistical descriptors for each set of charges (raw data) resulted from a single job, identified by EEM parameter set, computation method, and total molecular charge. Contents depend on the grouping and property menus above.",
Compare
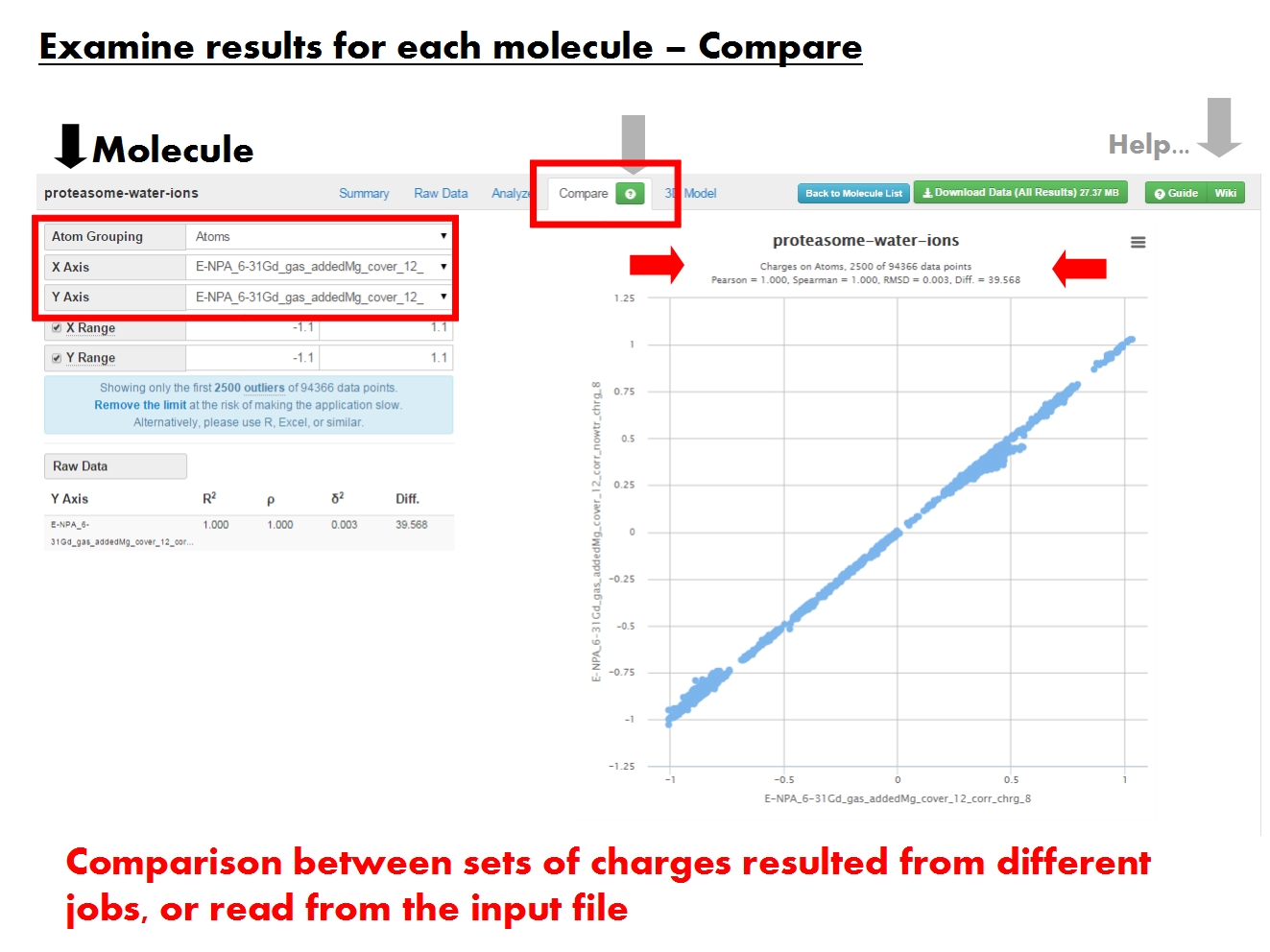
"ACC allows to compare sets of charges at more levels of resolution, in graphical and tabular form. These sets of charges may have resulted from different jobs, or were read from the input file.");
addStep("details-correlation-plot", //graph
"
Statistical comparison between two sets of charges (X and Y axes) on groups of atoms. For example, comparison between formal residue charges given in the input file, and residue charges as computed by EEM. Content of the graph dictated by the menus on the left.
",
"left");
addStep("charges-corr-controls", //AG, XY menus and outlier warning
"Dictate contents of the graph to the right, and of the Raw Data table below.",
"right");
addStep("charges-corr-ag", //atom grouping and menu
"Atom Grouping gives the level of resolution. Pick Atoms to compare atomic charges, or Residues to compare residue charges (sum of atomic charges for each residue). Residue-based statistics available only if residue annotations present in input file.",
"right");
addStep("charges-corr-xy", //X,Y and menus (not the ranges)
"Pick the sets of charges to compare. Each set of charges was produced by a single job (identified by EEM parameter set, computation method and total molecular charge), or was present in the input file (identified by the input file name and marked by ref).",
"right");
addStep("charges-corr-range", //X,Y ranges (entire lines)
"If you find the default XY ranges unsuitable for a certain dataset, you may adjust them manually.",
"right");
addStep("charges-corr-warn", //outliers warning
"
The number of data points depends on Atom Grouping. Comparison statistics computed using the entire datasets, but only 2500 data points display by default. Generally doesn't cause problems even when displaying 10000 data points. If in trouble, just reload the page.
" + "
Since outliers are displayed first, the graph might suggest the two charge sets are more dissimilar than the comparison statistics say.
",
"right");
addStep("charges-corr-raw", //Raw data and menu and table
"Tabular representation of the statistical comparison between a specific set of charges (raw data) and all other charge sets available for this molecule. Contents depend on the grouping menu above.",
"right");
3D Model
"ACC allows a few basic charge visualization options to aid in interpreting the results of the atomic charge calculation. You may need to update your browser and driver version.");
addStep("charges-3d-all-controls", //everything on the left side, but not the Update button
"Dictate contents of the visualization window on the right. 3D visualization is generally memory consuming, so you should consider the settings before attempting to visualize the 3D model of your molecule.",
"right");
addStep("details-3d-big-warning", //size warning
"
For large biomolecular complexes, start with lower levels of display detail, and work your way up if you feel unsatisfied with the quality of the display. If in trouble, just reload the page.
" + "
To reduce the complexity you may also choose to display the model based on residue positions (rather than atom positions). Only relevant if input file contains residue annotations.
",
"right");
addStep("charges-3d-update", //Update button
"Make sure your visualization settings are suitable, and click Update to display the 3D model of your molecule colored according to charges. Use Update whenever you modify the visualization options.",
"top");
addStep("details-3d-model", //visualizer window
"Visualization window, displaying a 3D model of your molecule, colored according to charges. Content depends on the visualization settings in the panel on the left. Mouse controls in the right bottom corner.",
"left");
addStep("charges-3d-ag", //atom grouping and menu
"Atom Grouping gives the level of resolution. Pick Atoms to build the 3D model based on atomic positions and color it by atomic charges. Pick Residues to build the 3D model based on residue positions and color it by residue charges (sum of atomic charges for each residue). Residue positions and charges available only if residue annotations present in input file. Requires Update to take effect.",
"right");
addStep("charges-3d-display", //display charges and menu
"Pick the sets of charges used in coloring the 3D model of your molecule. Each set of charges was produced by a single job (identified by EEM parameter set, computation method and total molecular charge), or was read from the input file (identified by the input file name and marked by ref).",
"right");
addStep("charges-3d-diff-wrap", //show differences and menu
"Instead of coloring by charges, it is sometimes useful to color by differences between two sets of charges. For example, to see how the presence of water changes the charge profile of the molecular surface.",
"right");
addStep("charges-3d-mode", //display mode and dependent menus
"
Several basic modes available for displaying the 3D model, depending on the Atom Grouping. Requires Update to take effect.
" + "
Each display mode has additional options you may wish to tweak. For example, display the protein using Cartoons, and the ligands using Balls and Sticks, but hide water molecules.
",
"right");
addStep("charges-3d-detail", //display detail and menu
"For large biomolecular complexes, start with lower detail. Requires Update to take effect.",
"right");
addStep("charges-3d-colors", //Color scale menus
"If you find the default coloring scheme unsuitable, you may adjust it manually.",
"right");
addStep("charges-3d-gap", //Value gap and menu
"Further adjustment to the coloring scheme allows to emphasize the visual difference in charge distribution in different areas of the molecule. Emphasis increases with Value Gap.",
"right");
addStep("charges-3d-default", //Set default parameters
"Reset to default visualization options. Might require Update to take effect.",
Return to the Table of contents.