ChargeCalculator:Job submission: Difference between revisions
No edit summary |
|||
| Line 33: | Line 33: | ||
If bond information is not explicitly included in the input file, '''ACC''' will attempt to compute this information based on the molecular structure. This algorithm may assign wrong bond information when interatomic distances vary significantly from the expected norms. This behavior may only affect calculations using EEM parameter sets which distinguish between atom types based on bond information. | If bond information is not explicitly included in the input file, '''ACC''' will attempt to compute this information based on the molecular structure. This algorithm may assign wrong bond information when interatomic distances vary significantly from the expected norms. This behavior may only affect calculations using EEM parameter sets which distinguish between atom types based on bond information. | ||
<br style="clear:both" /> | <br style="clear:both" /> | ||
Revision as of 06:56, 29 November 2014
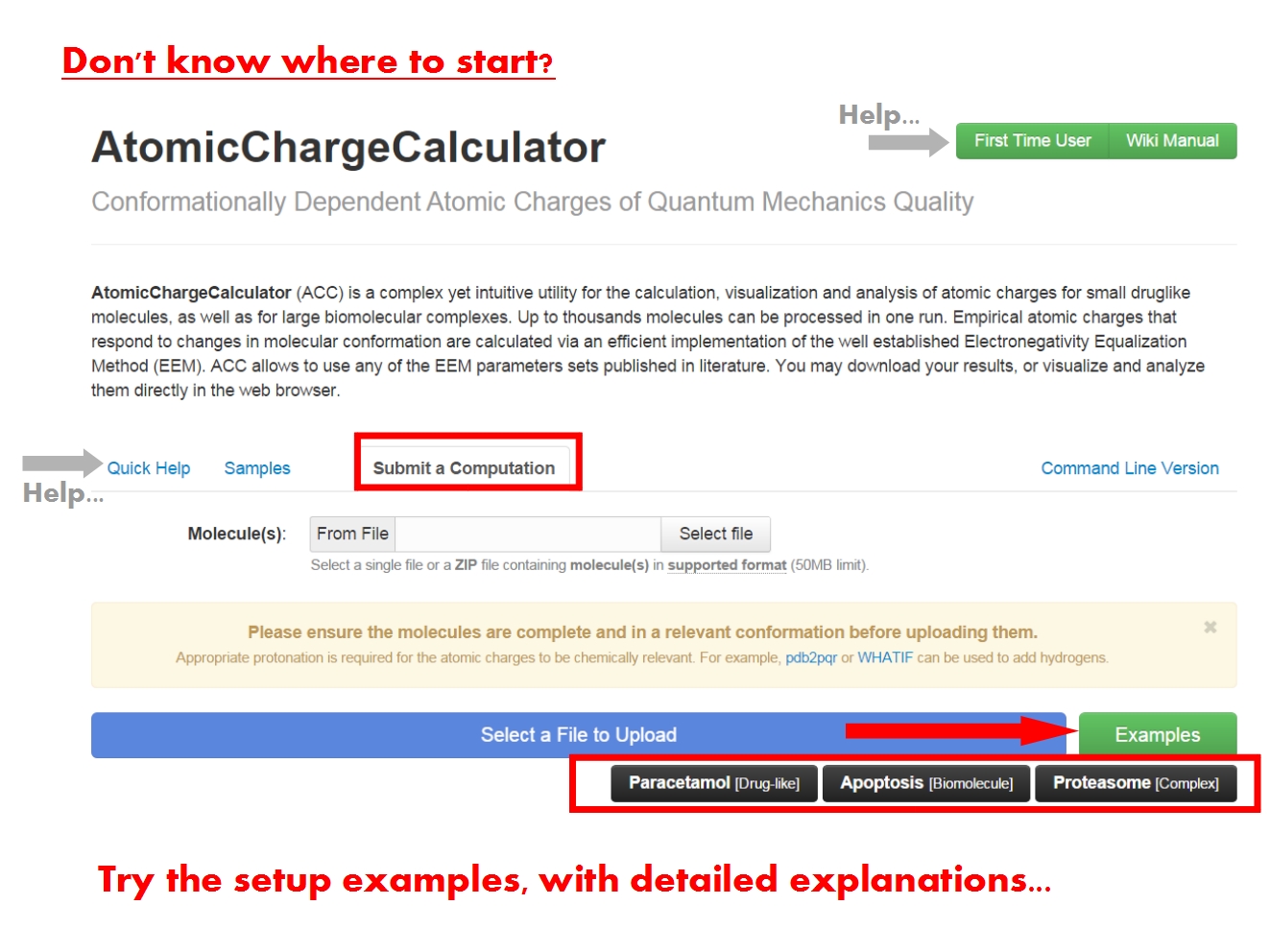
The ACC submission page contains a brief description of ACC, and is further organized into several tabs. Since this is your first contact with ACC, the support tabs Quick Help and Samples help you get started, with basic information and examples of ACC use cases.
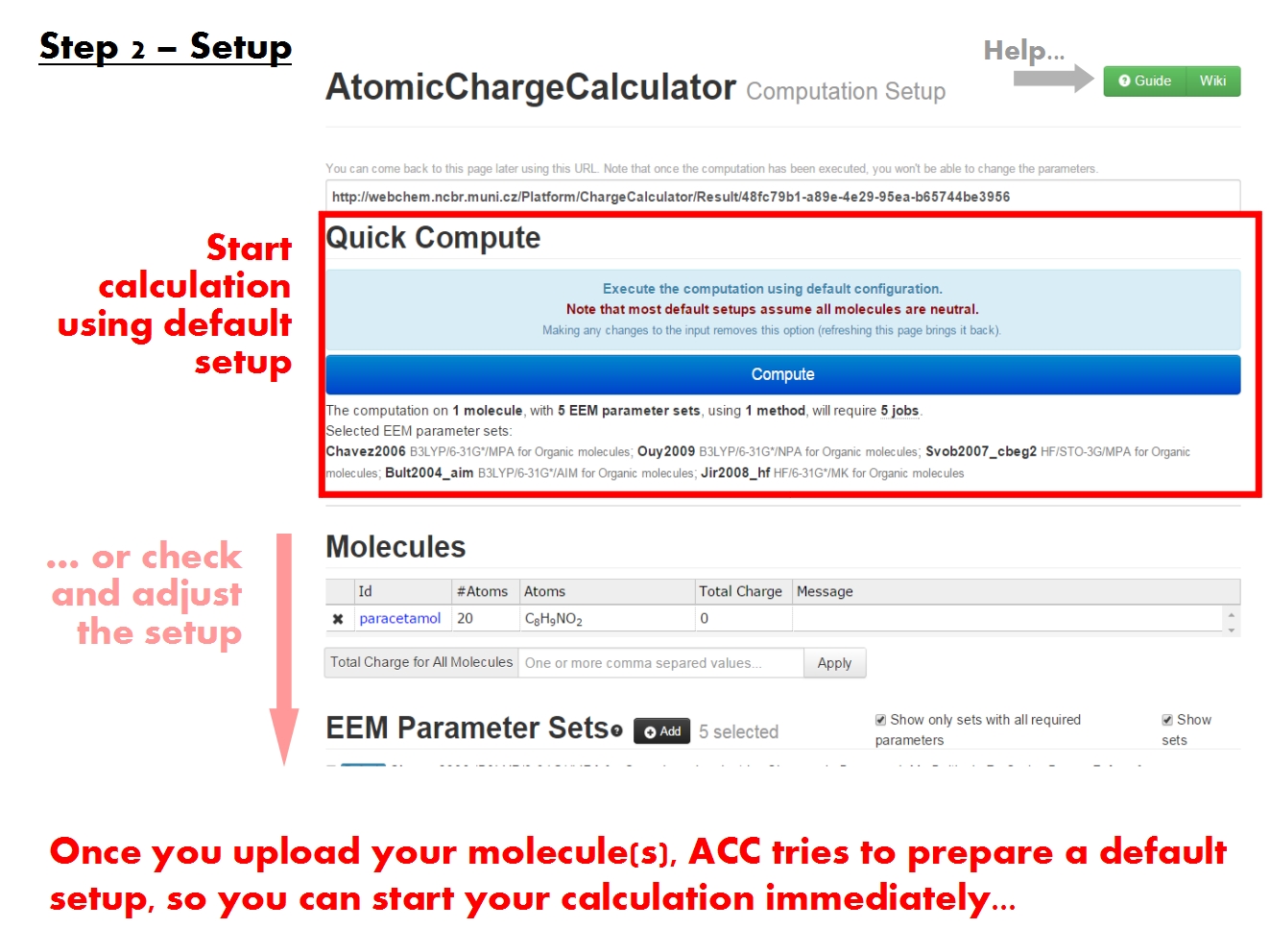
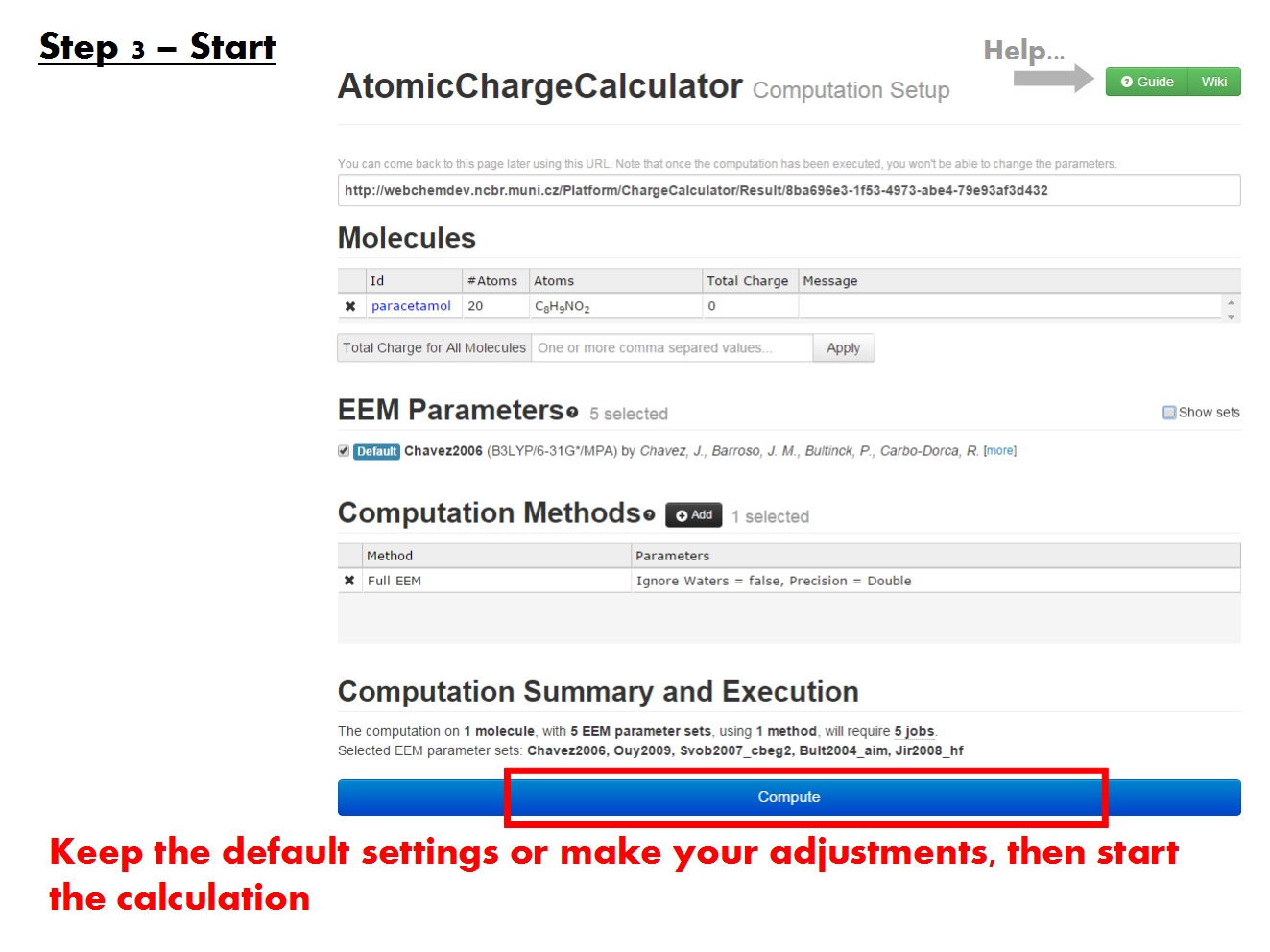
You may submit an atomic charge calculation in 3 easy steps, namely: upload your molecule(s), adjust the calculation setup, and finally start the calculation.
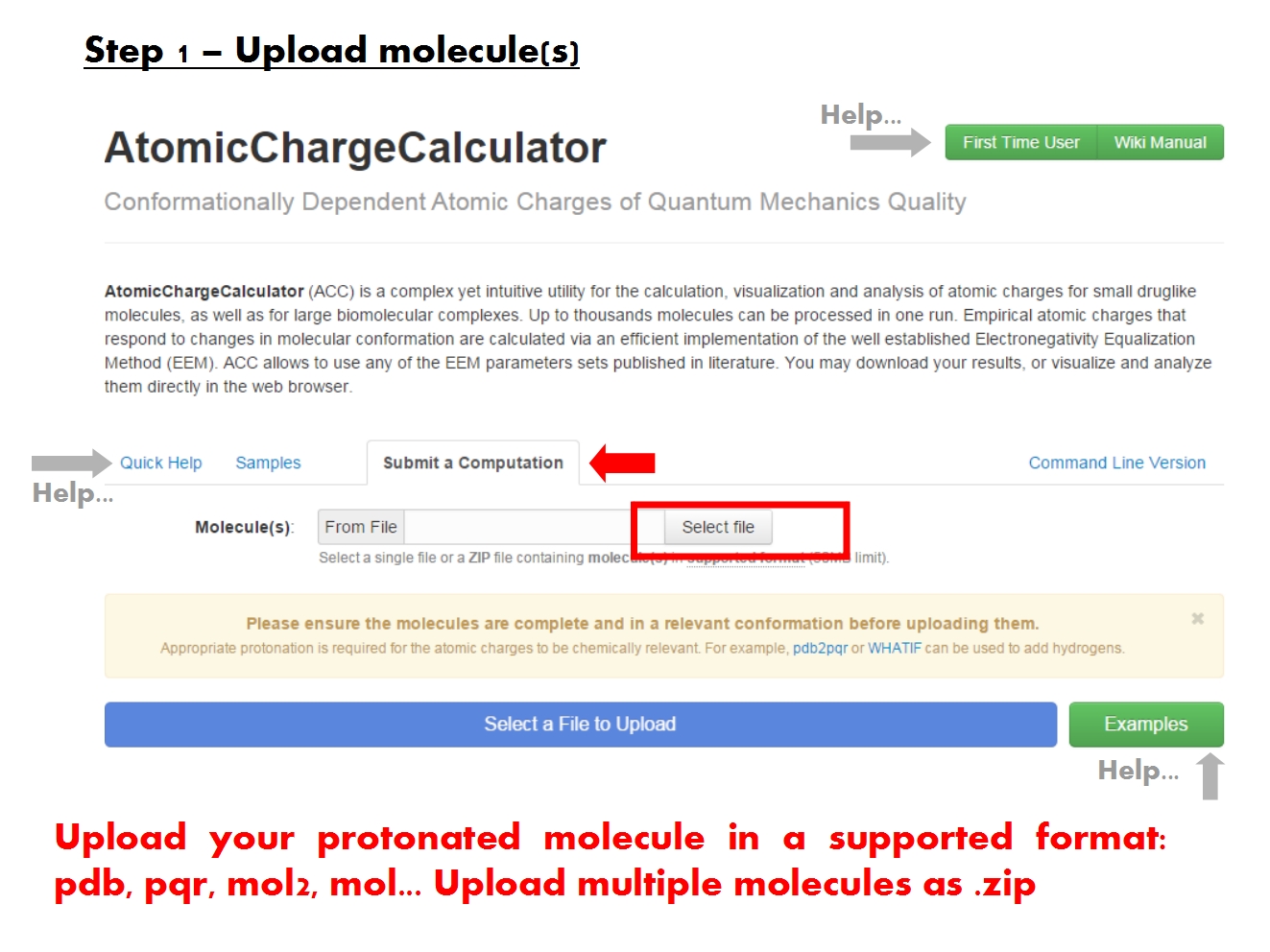
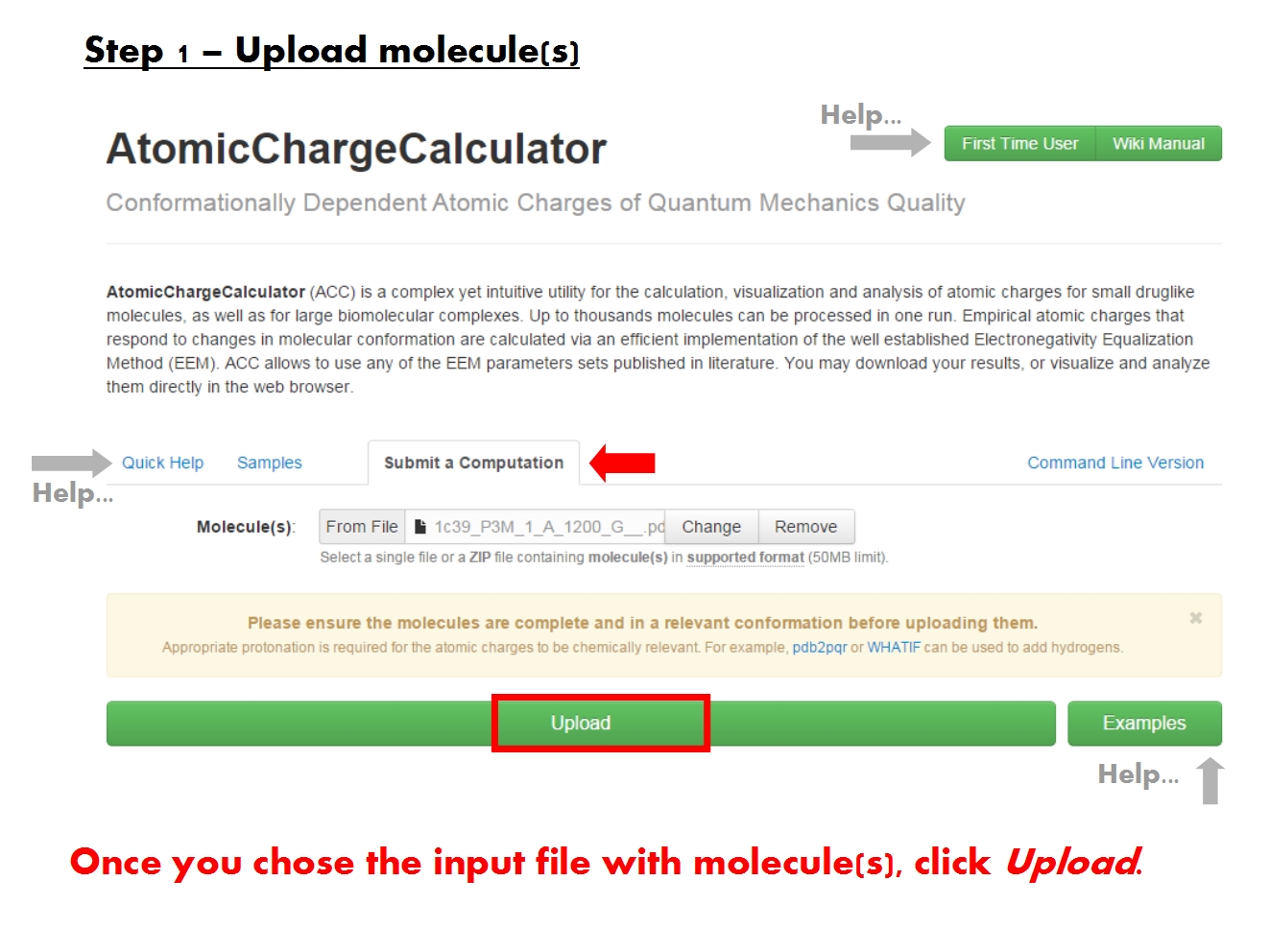
Upload molecules
ACC is able to read the molecular structure and charge information from the most common file formats. Nonetheless, because it was designed to handle molecules of all kinds and size, ACC generally requires that the input files follow the formal guidelines established for each format.
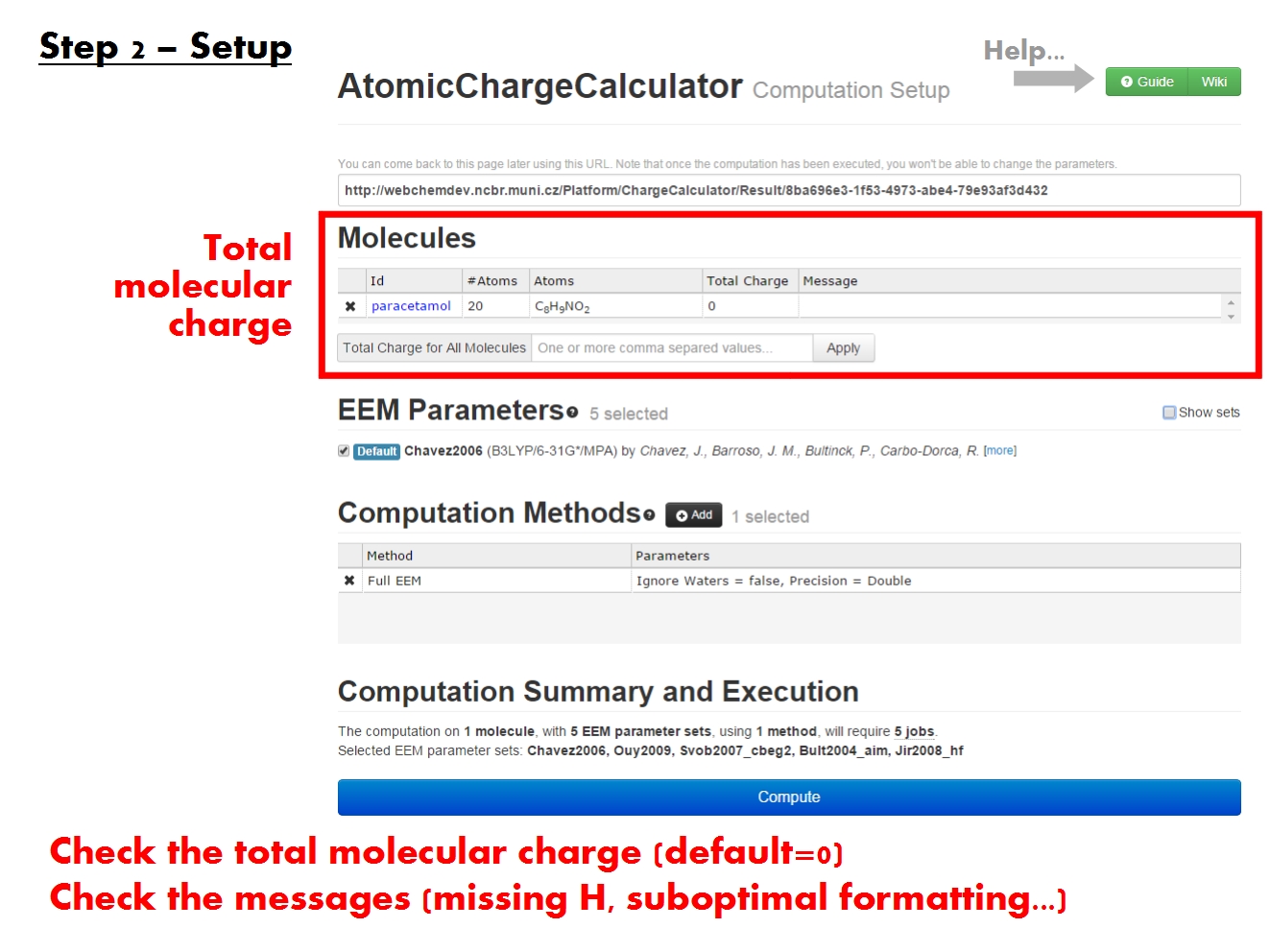
In order to produce chemically relevant atomic charges using EEM, it is necessary that the structure of the molecule be complete. No crucial parts should be missing. If parts of the structure are missing, appropriate cappings should be included. All protons should be present according to the relevant protonation state. Since ACC does not currently include functionality for editing the molecular structure, you must address these issues prior to uploading the molecule into ACC.
ACC does check valences, and produces a missing H warning if they are not satisfied. Despite the warning though, ACC allows to proceed with the charge calculation step, as it might not always be possible to obtain a perfect structure (e.g., when working with low resolution structures of extremely large complexes). The results from such calculations may not have chemical meaning in their absolute values, but they can be very useful when comparing sets of charges (open vs closed conformation, free vs bound state, etc.).
Input files generally contain atom type information. Many different atom type schemes are used in different modeling projects. Moreover, many times the output is not even standardized between different applications implementing the same atom type scheme. ACC attempts to be a general utility, and currently implements only the detection of chemical elements. If the atom types in the input file differ from chemical elements, ACC will report them as unknown chemical elements, and these atoms will be skipped during the EEM calculation (they will not contribute to the EEM matrix ). A similar problem will arise if the atom type information is not found at the expected place in the file. In the future, a more complex parsing algorithm may be implemented in ACC in order to cover the most common atom type schemes (e.g., AMBER, OPLS, etc.). Currently, the atom type parsing problem can be worked around either by uploading input files which adhere to the formal guidelines for their respective formats and contain atom types according to chemical elements, or by creating an EEM parameter set with special parameters for those atom types which ACC finds problematic.
If the chain ID is not explicitly included in the input file, but the molecule contains multiple chains with overlapping residue serial numbers, the results will not be meaningful for the affected residues, and possibly even in the vicinity of these residues. ACC provides check chain ID warnings both before and after the computation if this problem is detected, so that the input file can be corrected.
If bond information is not explicitly included in the input file, ACC will attempt to compute this information based on the molecular structure. This algorithm may assign wrong bond information when interatomic distances vary significantly from the expected norms. This behavior may only affect calculations using EEM parameter sets which distinguish between atom types based on bond information.
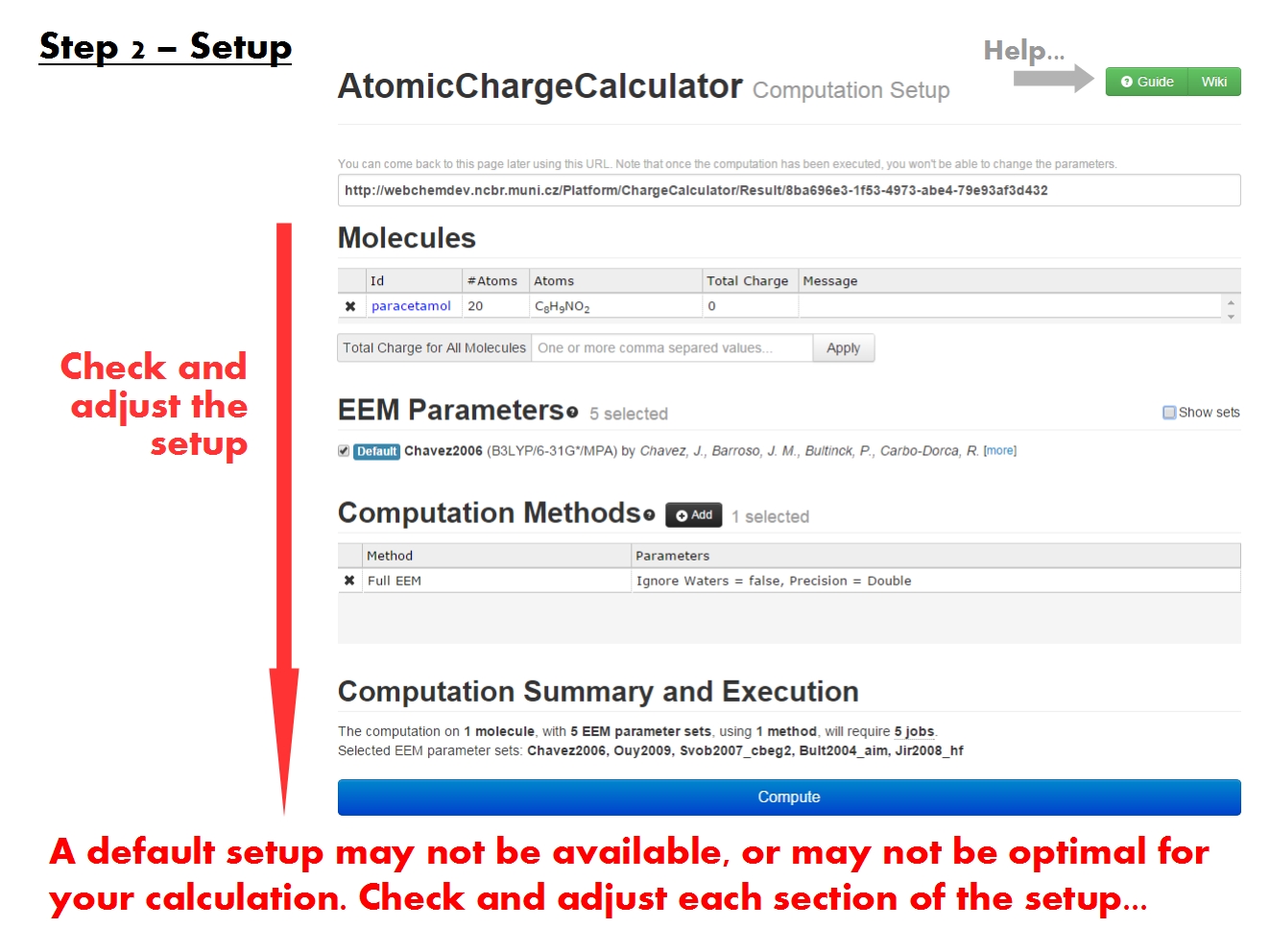
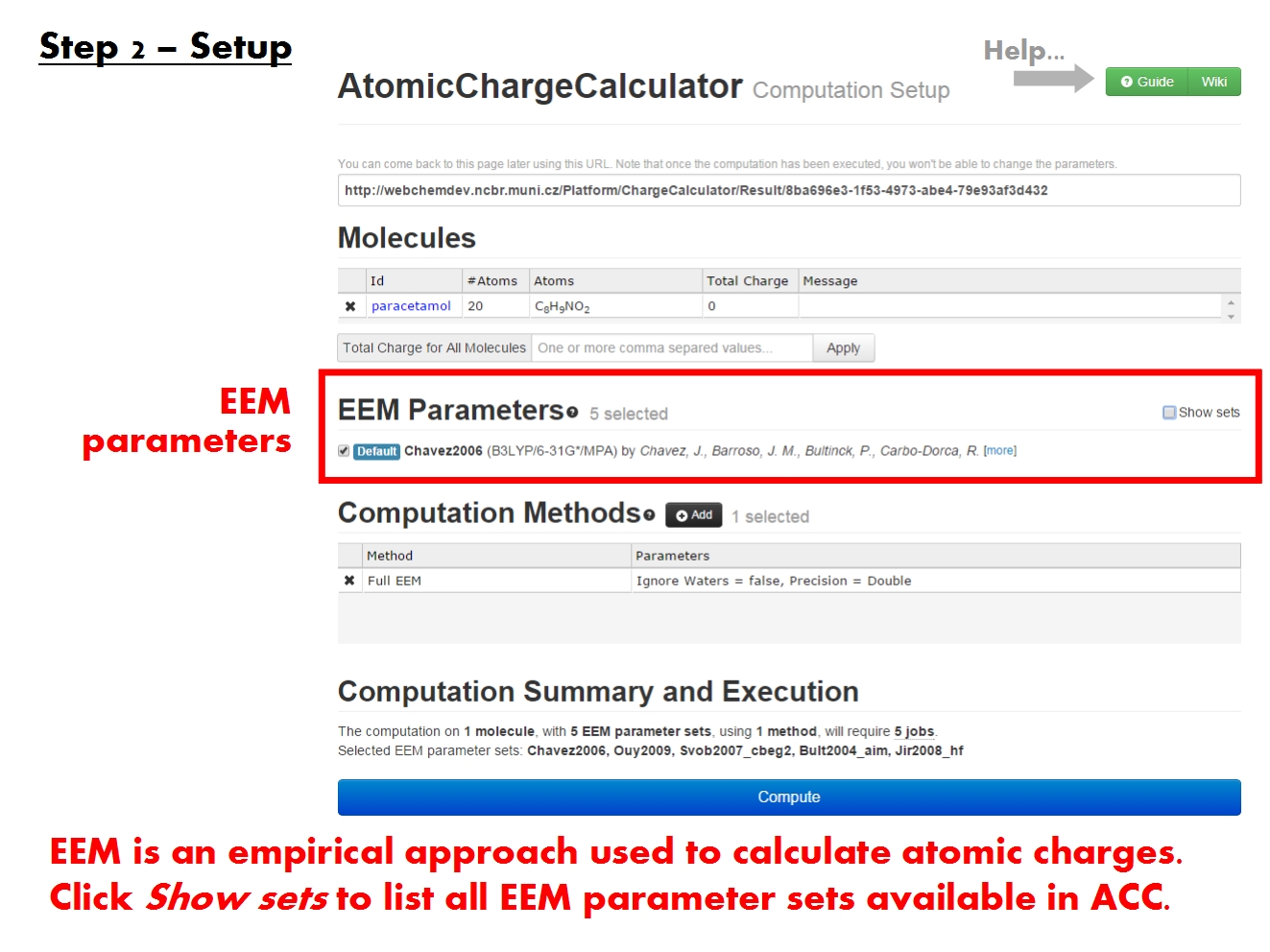
Setup computation
ValidatorDB contains validation results for the entire Protein Data Bank. You are probably interested only in a subset of these (e.g., the validation results for a specific inhibitor which appears in the PDB entry you plan to use further in your simulations). The synopsis page allows to access specific validation reports via the Search tab..................
|
|
| Caption for download. | Caption for molecules. |
The validation results are accessible in tabular and graphical form via several tabs on the specifics page, namely Overview, Summary, Details and Processing Warnings. These are described in detail in their respective sections below. It is important to note that the specifics page contains annotation-based validation reports. This means that if the molecules you selected share the same annotation (3-letter code), all results will be included in a single report. However, you may have retrieved validation results for molecules with different annotations, as is the case with PDB entries containing many different ligands for instance. In this case, the validation results will be organized according to annotation. Specifically, there will be as many reports as there are annotations. All tabs on the specifics page discuss one report at a time, and allow easy navigation between these reports. The only exception is the Overview tab, which presents the total statistics for all validated molecules, and not per annotation................
You may use the JSON download button at the top right corner of the page in order to download the complete validation reports and perform any additional analyses on your own.
Set total charge
The validation reports available in the Summary tab of the specifics page are organized by annotation (3-letter code). You can easily navigate between reports by clicking on the annotation of interest in the bar under the tab names. Each report consists of information about the model and validated molecules corresponding to that particular annotation.
First, the model from wwPDB CCD is briefly described by name, molecular formula, 2D structure, etc. Next, a recapitulation of all molecules and their respective PDB entries is also included, followed by a table with results of the main validation analyses (completeness, chirality, advanced). For each validation analysis, statistics are provided for all problematic atoms. Note that here the statistics cover only the molecules sharing this annotation (3-letter code).
The first table with validation results is organized into two main sections, referring to incomplete (Missing Atoms or Rings) and complete structures. respectively. The section with complete structures is further organized into subsections based on the results of the chirality and advanced analyses. Specifically, a general distinction is made between complete structures with correct chirality and with wrong chirality. Warnings like foreign atom and substitutions are also included.
Subsequent tables highlight problematic atoms and provide useful statistics, in order to better localize the problems in the structures.
If you click on any validation issue (e.g., missing rings, wrong chirality on a specific atom), you will be redirected to the Details tab where only the molecules with those specific issues are listed.
Results can be interpreted using the information in the Database contents. Tool tips are available for each element of the table headers. The results for different types of validation analyses are labeled using the unified color scheme: complete structure and correct chirality, incomplete structure, wrong chirality, warning.
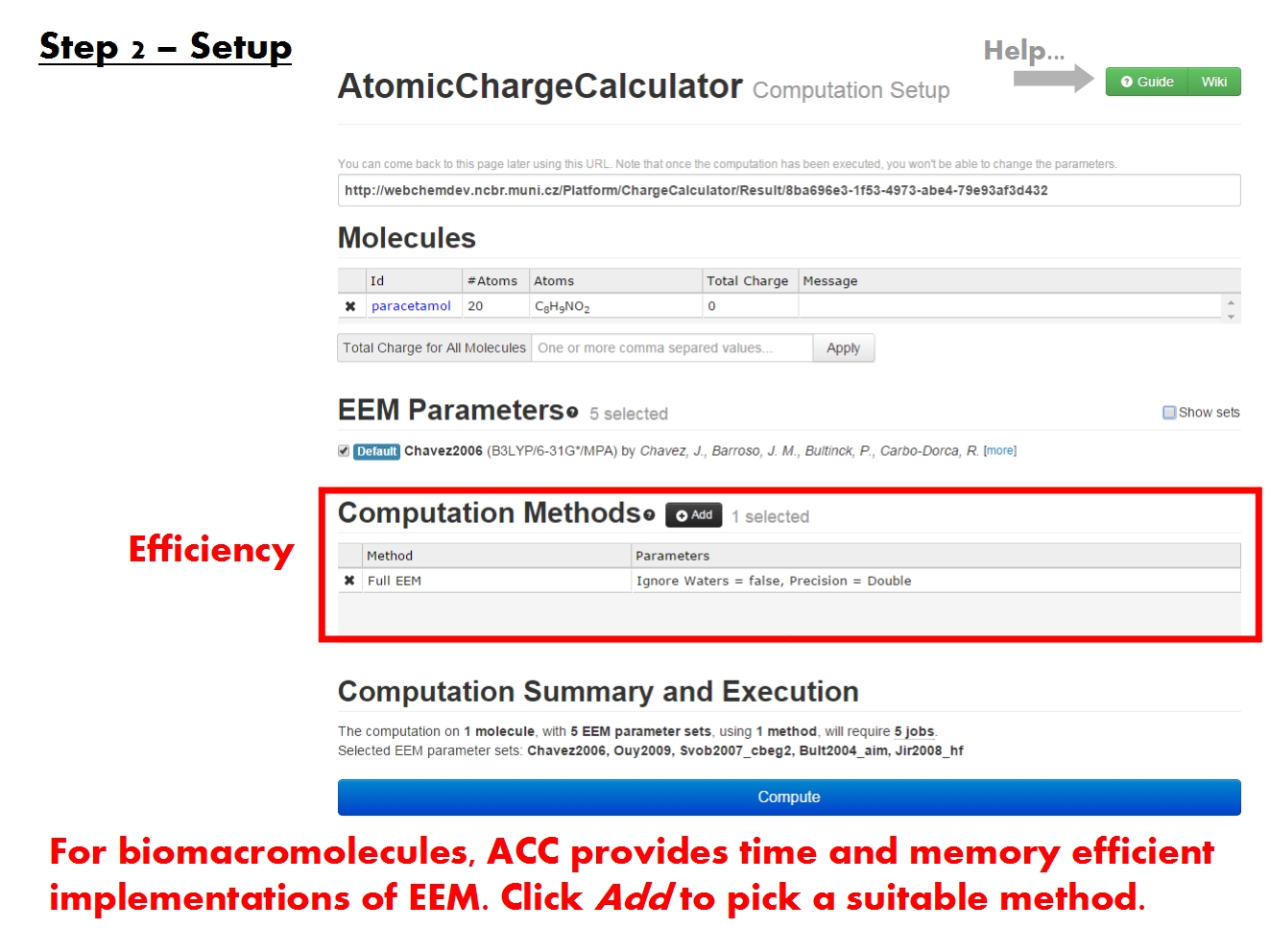
Pick EEM parameters
The single molecule report offers a complete image of the validation results for each particular molecule. It is accessible via the Details tab on the specifics page, by clicking on the Id (unique identifier containing the PDB ID and a serial number) of the molecule of interest. As you open this report, the molecule Id will appear in the top left corner of the report.
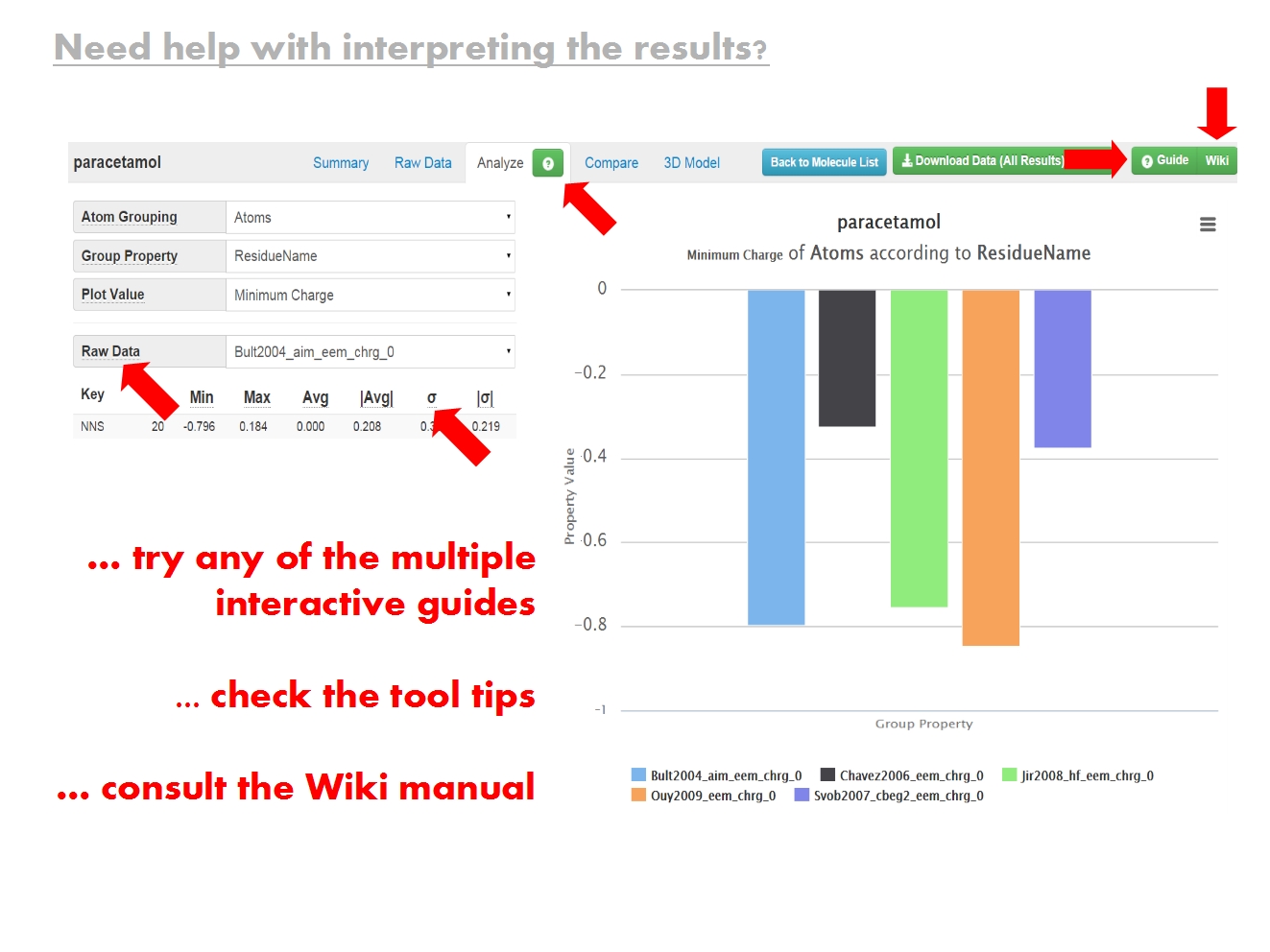
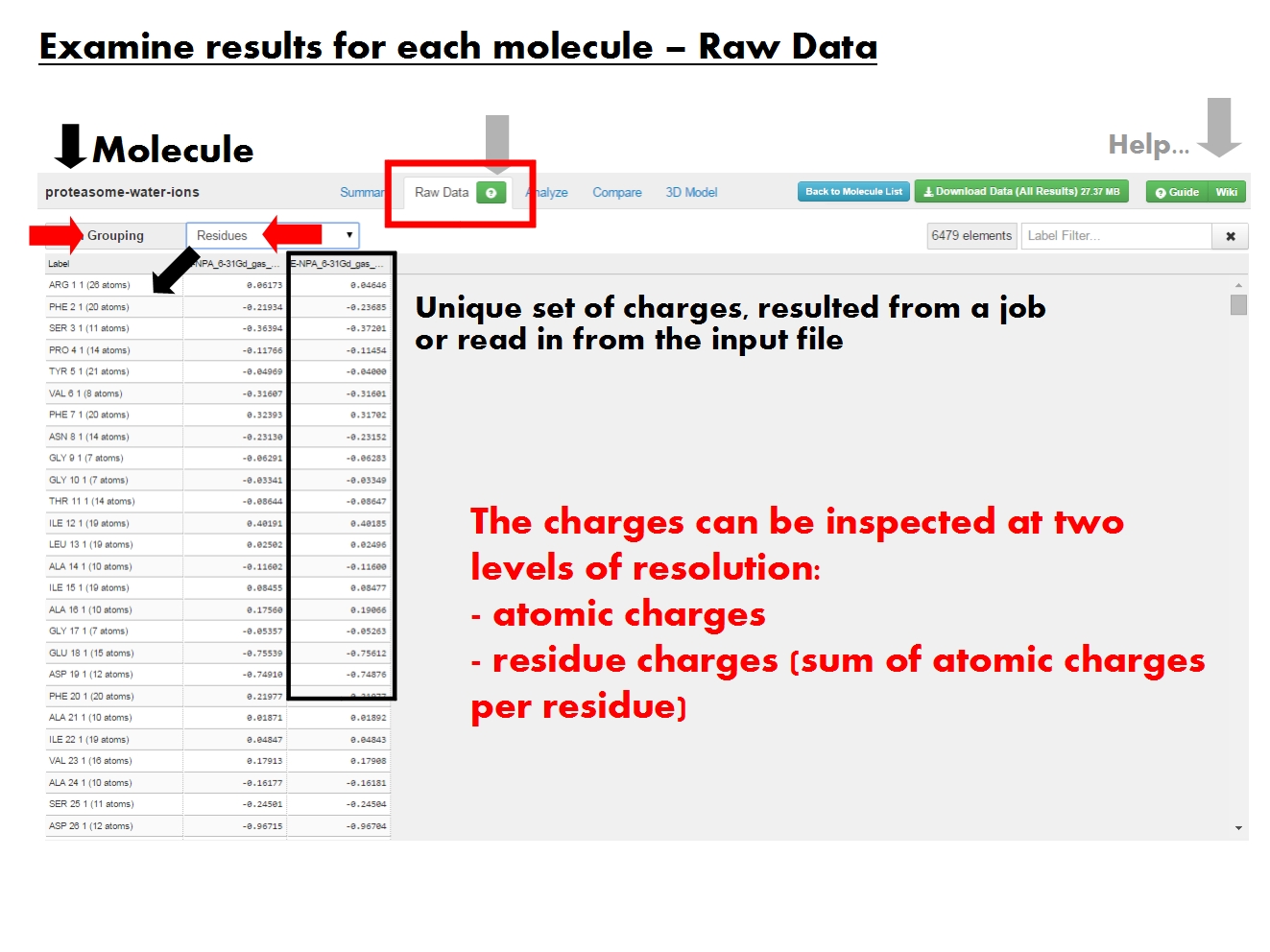
Analyze
Whereas the Summary tab of the synopsis page provides statistics, the Details tab allows you to filter the validation results according to the results of the various validation analyses. Note that you may also access relevant information in the Details tab by clicking on a specific validation issue in any of the tables in the Summary tab.
Pick computation method
The single molecule report offers a complete image of the validation results for each particular molecule. It is accessible via the Details tab on the specifics page, by clicking on the Id (unique identifier containing the PDB ID and a serial number) of the molecule of interest. As you open this report, the molecule Id will appear in the top left corner of the report.
Start computation
The specifics page may also contain the separate tab Processing warnings, which lists unusual circumstances encountered during the validation of each molecule. Sometimes the PDB entries contain information that is ambiguous, conflicting or which deviates strongly from the expected reference.
Some Processing warnings are issues that may cause incorrect validation, such as two molecules being too close together in the 3D space, or some atoms being too far (disconnected) from the rest of the molecule. Such structures are generally marked as degenerate in the completeness analyses. Other processing warnings are only meant to inform of which course of action was taken during validation (e.g., which conformer was validated if several conformers of that molecule were present in the original PDB entry).
The drop down menu at the top of the page helps filter the warnings for molecules with a specific annotation (3-letter code).
Return to the Table of contents.