ValidatorDB:Database contents: Difference between revisions
No edit summary |
|||
| Line 18: | Line 18: | ||
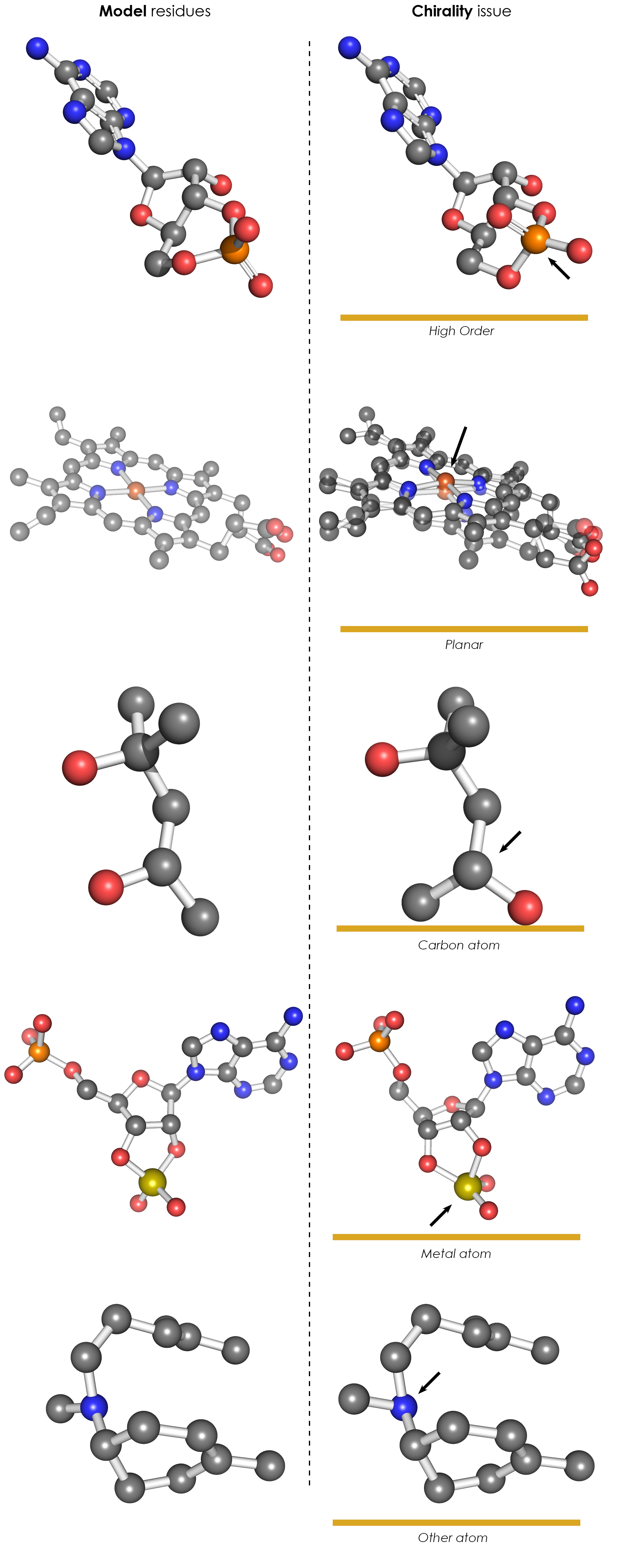
[[File:VDB chirality.png|thumb|right|400px| Complete structures can contain chirality issues on a variety of different atom types, here all possible cases occuring in '''ValidatorDB''' are illustrated.]] | [[File:VDB chirality.png|thumb|right|400px| Complete structures can contain chirality issues on a variety of different atom types, here all possible cases occuring in '''ValidatorDB''' are illustrated.]] | ||
If no issue was found during the ''completeness'' analyses, the validated molecule is marked as ''complete'', because it contains all the atoms which are present in the model. ''Chirality'' is further evaluated only for these molecules with ''complete structure'', because the absence of some atoms can make it difficult to check the chirality of the remaining atoms. | |||
==Wrong chirality== | ==Wrong chirality== | ||
'''ValidatorDB''' presents the results of several kinds of ''chirality'' analyses. If an issue is found, the following types of results are reported: | |||
** ''' | ** '''Wrong chirality (Carbon)''': the chirality error was found on an sp3 hybridized carbon atom. | ||
** '''Planar | ** '''Wrong chirality (Planar)''': the chirality error was found on a planar chiral center. Because of their spacial distribution, planar chiral centers are very sensitive even to small perturbations in the position of the substituents. Therefore, some of the errors reported here might not be significant. | ||
** '''Metal | ** '''Wrong chirality (Metal)''': the chirality error was found on a metal chiral center. | ||
** '''High order | ** '''Wrong chirality (High order)''': the chirality error was detected on a chiral atom (e.g., phosphorus) where at least one substituent is bound by a bond of higher order. | ||
** '''Other''': Additional chirality issue which could not be | ** '''Other''': Additional chirality issue which could not be listed in any of the above categories (e.g., chirality issues on nitrogen atoms). These appear rarely in the Protein Data Bank (<0.5% of molecules). | ||
If any issue is detected on any atom during any ''chirality'' analysis, the molecule is marked as having ''wrong chirality'', because there is at least one atom in the validated molecule with different chirality than the corresponding atom in the model. If more atoms in a molecule exhibit the same type of chirality issue, the validated molecule is counted only once in each type of statistics (e.g., if 3 carbon atoms in one glucose molecule have wrong chirality, this glucose molecule will contribute only once to the statistics on "wrong chirality (carbon)"). If a validated molecule exhibits issues in more than one type of ''chirality'' analysis, it will be counted once in each corresponding statistics (e.g., heme molecules may appear both in the statistics for ''wrong chirality (planar)'', and for the ''wrong chirality (metal)''). | |||
==Correct chirality== | ==Correct chirality== | ||
If no issues are detected during the '' | If no issues are detected during the ''chirality'' analyses, the validated molecule is marked as having '''correct chirality''', because it contains all atoms present in the model, and each of these atoms has the same chirality in the validated molecule and in the model. | ||
Some types of chirality errors do not constitute real issues, but are artifacts of the automated chirality determination procedure. Specifically, an error in planar chirality may just mean that the chiral atom is situated slightly above or below the plane compared to its equivalent in the model from wwPDB CCD. Further, an error in high order chirality often marks the involvement of phosphate O atoms in salt or ester formation, or merely a different PDB format identification of phosphate O atoms of the validated molecule compared to the model. Therefore, if the validated molecule is found to have planar or high order chirality errors, but no other type of chirality issues, the molecule is marked as having '''correct chirality (tolerant)'''. | |||
<br style="clear:both" /> | <br style="clear:both" /> | ||
Revision as of 10:11, 8 September 2014
Please make sure to go through the terms and principles governing ValidatorDB before you read this page.
For each molecule, the validation report contains several types of results, based on how the molecule fared during the various validation analyses . The evaluation relies on comparing all atoms and bonds in each validated molecule to those in the corresponding model. ValidatorDB reports correct structures, as well as all potential issues found during validation, namely structures that are wrong either because they are incomplete (missing atoms or rings), or because the chirality of some atoms is incorrect. If no issues are found during the completeness and chirality analyses, the molecule is marked as having complete structure and correct chirality. The results of the advanced analyses are reported as warnings. Additionally, unusual circumstances encountered during validation are reported separately as processing warnings.
In ValidatorDB, the results for different types of validation analyses are labeled using the unified color scheme: complete structure and correct chirality, incomplete structure, wrong chirality, warning.
Incomplete structures
Validated molecules exhibiting an error in at least one of the Completeness analyses are denoted as incomplete, whereas the remaining molecules are reported as complete. Incomplete structures refer to molecules which lack some atoms, or whose structure is significantly distorted in comparison to the model:
- Missing atoms: An atom in the model has no corresponding atom in the validated molecule.
- Missing rings: At least one missing atom originates from cycles (rings). The formal distinction between ring atoms and non-ring atoms (simply denoted as atoms) is meant to allow a quick localization of potential issues in molecules containing rings, especially where atom identifiers are not useful.
- Degenerate: The molecules could not be validated due to their distorted structure, i.e. significant discrepancies between the atoms and inter-atomic bonds in the validated molecule and in the model. This generally happens when residues are overlapping in the 3D space, or some atoms appear disconnected from the rest of the structure.
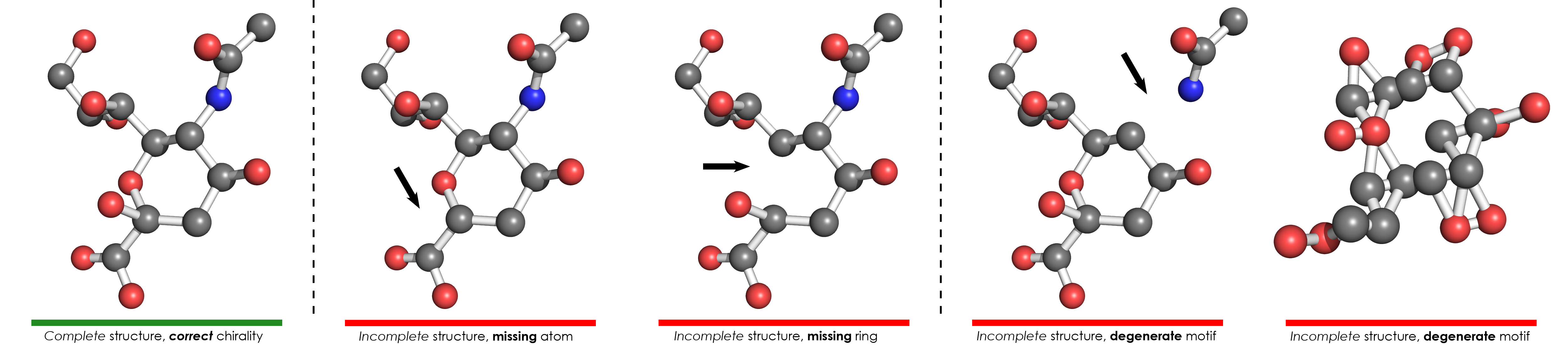
Complete structures
If no issue was found during the completeness analyses, the validated molecule is marked as complete, because it contains all the atoms which are present in the model. Chirality is further evaluated only for these molecules with complete structure, because the absence of some atoms can make it difficult to check the chirality of the remaining atoms.
Wrong chirality
ValidatorDB presents the results of several kinds of chirality analyses. If an issue is found, the following types of results are reported:
- Wrong chirality (Carbon): the chirality error was found on an sp3 hybridized carbon atom.
- Wrong chirality (Planar): the chirality error was found on a planar chiral center. Because of their spacial distribution, planar chiral centers are very sensitive even to small perturbations in the position of the substituents. Therefore, some of the errors reported here might not be significant.
- Wrong chirality (Metal): the chirality error was found on a metal chiral center.
- Wrong chirality (High order): the chirality error was detected on a chiral atom (e.g., phosphorus) where at least one substituent is bound by a bond of higher order.
- Other: Additional chirality issue which could not be listed in any of the above categories (e.g., chirality issues on nitrogen atoms). These appear rarely in the Protein Data Bank (<0.5% of molecules).
If any issue is detected on any atom during any chirality analysis, the molecule is marked as having wrong chirality, because there is at least one atom in the validated molecule with different chirality than the corresponding atom in the model. If more atoms in a molecule exhibit the same type of chirality issue, the validated molecule is counted only once in each type of statistics (e.g., if 3 carbon atoms in one glucose molecule have wrong chirality, this glucose molecule will contribute only once to the statistics on "wrong chirality (carbon)"). If a validated molecule exhibits issues in more than one type of chirality analysis, it will be counted once in each corresponding statistics (e.g., heme molecules may appear both in the statistics for wrong chirality (planar), and for the wrong chirality (metal)).
Correct chirality
If no issues are detected during the chirality analyses, the validated molecule is marked as having correct chirality, because it contains all atoms present in the model, and each of these atoms has the same chirality in the validated molecule and in the model.
Some types of chirality errors do not constitute real issues, but are artifacts of the automated chirality determination procedure. Specifically, an error in planar chirality may just mean that the chiral atom is situated slightly above or below the plane compared to its equivalent in the model from wwPDB CCD. Further, an error in high order chirality often marks the involvement of phosphate O atoms in salt or ester formation, or merely a different PDB format identification of phosphate O atoms of the validated molecule compared to the model. Therefore, if the validated molecule is found to have planar or high order chirality errors, but no other type of chirality issues, the molecule is marked as having correct chirality (tolerant).
Warnings
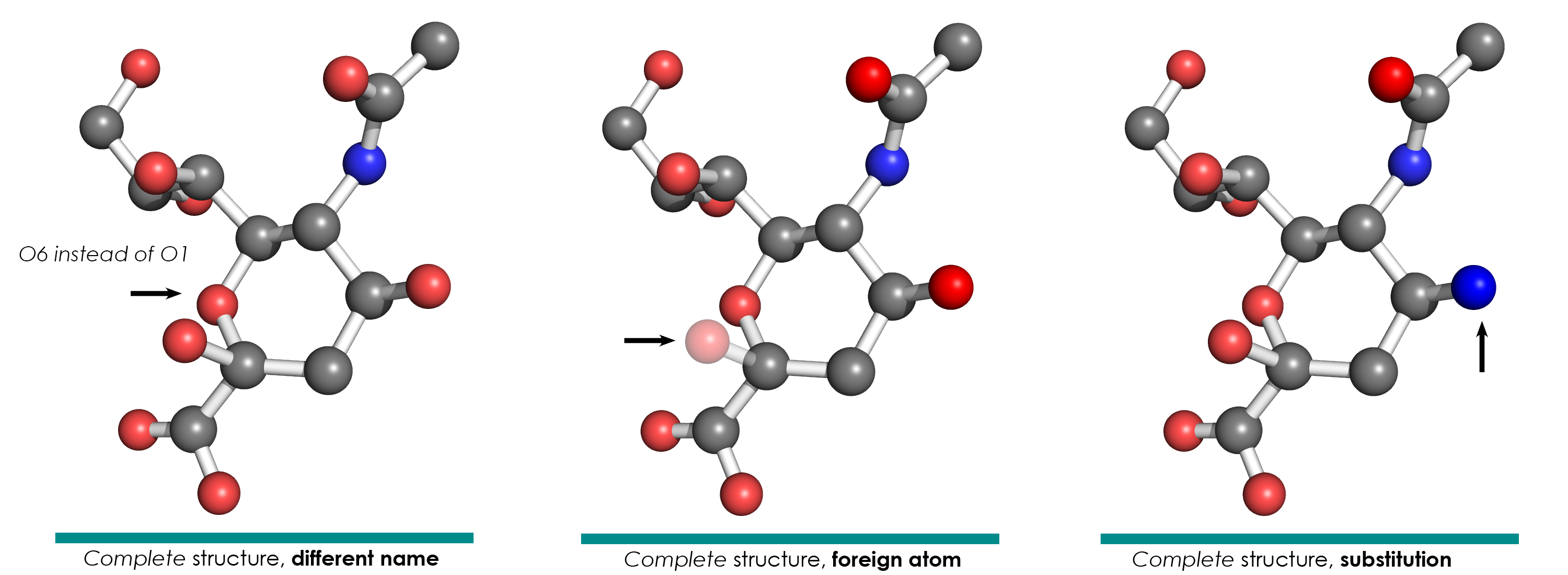
- Substitution: An atom from the validated motif is of a different chemical element than the corresponding atom in the model residue (e.g. O mapped to N). This happens often at linkage sites.
- Different naming: An atom from the validated motif has a different PDB atom name than the corresponding atom from the model residue (e.g. the C1 atom mapped to the C7 atom). This happens often when the original PDB files were produced by different software.
- Foreign atom: An atom from the model residue was mapped to an atom from outside the validated residue (i.e. from its surroundings).
- Alternate conformations: In the original PDB file, the validated residue contains atoms which were given in conformations (i.e., most probably different rotamers). Only the first rotamer was considered during validation.
- Zero model RMSD: The superimposition between the model residue and the validated motif has a root mean square deviation of zero, i.e., the validated motif is identical to the model residue used as reference.
While the results of the Advanced analyses have no bearing over the chemical soundness of the validated molecules, they indicate that further, especially automated processing of these structures can be very problematic. Comparison between the structures of molecules with the same annotation (3-letter code) from different PDB entries might even be impossible in the presence of a substitution, as the corresponding atoms have different chemical elements. PDB atom names cannot be used straightforwardly, since even element symbols can differ and atoms can be formally included in neighboring residues.
Processing warnings
Furthermore, for each validation run, there is Processing warnings tab available, where one can find warnings about the processed structures. Typically, there is an information about skipped alternate conformation residues, wrong CONNECT records from the parent PDB file, processed models (in case the parent PDB is composed of multiple models) etc. Finally, as a general rule, in the validation interface, errors are marked in red (missing atoms) or dark yellow (wrong chirality), correct structures in green, and warnings in cyan.