SecStrAnnotator:Analysis: Difference between revisions
No edit summary |
No edit summary |
||
| Line 24: | Line 24: | ||
|} | |} | ||
<div align="left" style="display: inline;"> | |||
[[File:SecStrAnnotator-cyp-sse-occurrence.png | 500px | Occurrence of particular SSEs in the whole set. ]] | |||
[[File:SecStrAnnotator-cyp-sse-occurrence-Bact-Euka.png | 500px | Occurrence of particular SSEs – comparison of bacterial and eukaryotic structures. ]] | |||
</div> | |||
===Length of SSEs=== | ===Length of SSEs=== | ||
Revision as of 12:19, 20 March 2020
SecStrAnnotator Suite provides scripts (Python, R, and bash) for batch annotation of the whole family and analysis of the annotation results. The main scripts are
- SecStrAPI_master.sh – this bash script used to prepare data for SecStrAPI also formats the annotations into tab-separated (TSV) files readable by R
- secondary_structure_anatomy.R – contains commands for reading the annotation results, generating plots, and performing some statistical test to compare eukaryotic and bacterial structures (or any two sets of structures)
Example case study: Cytochromes P450
Data
For the Cytochrome P450 family, structures of 1775 protein domains are available, located in 953 PDB entries. The analysis was performed on a non-redundant subset containing 175 protein domains.
Occurrence of SSEs
The occurrence describes in what percentage of the structures a particular SSE is present.
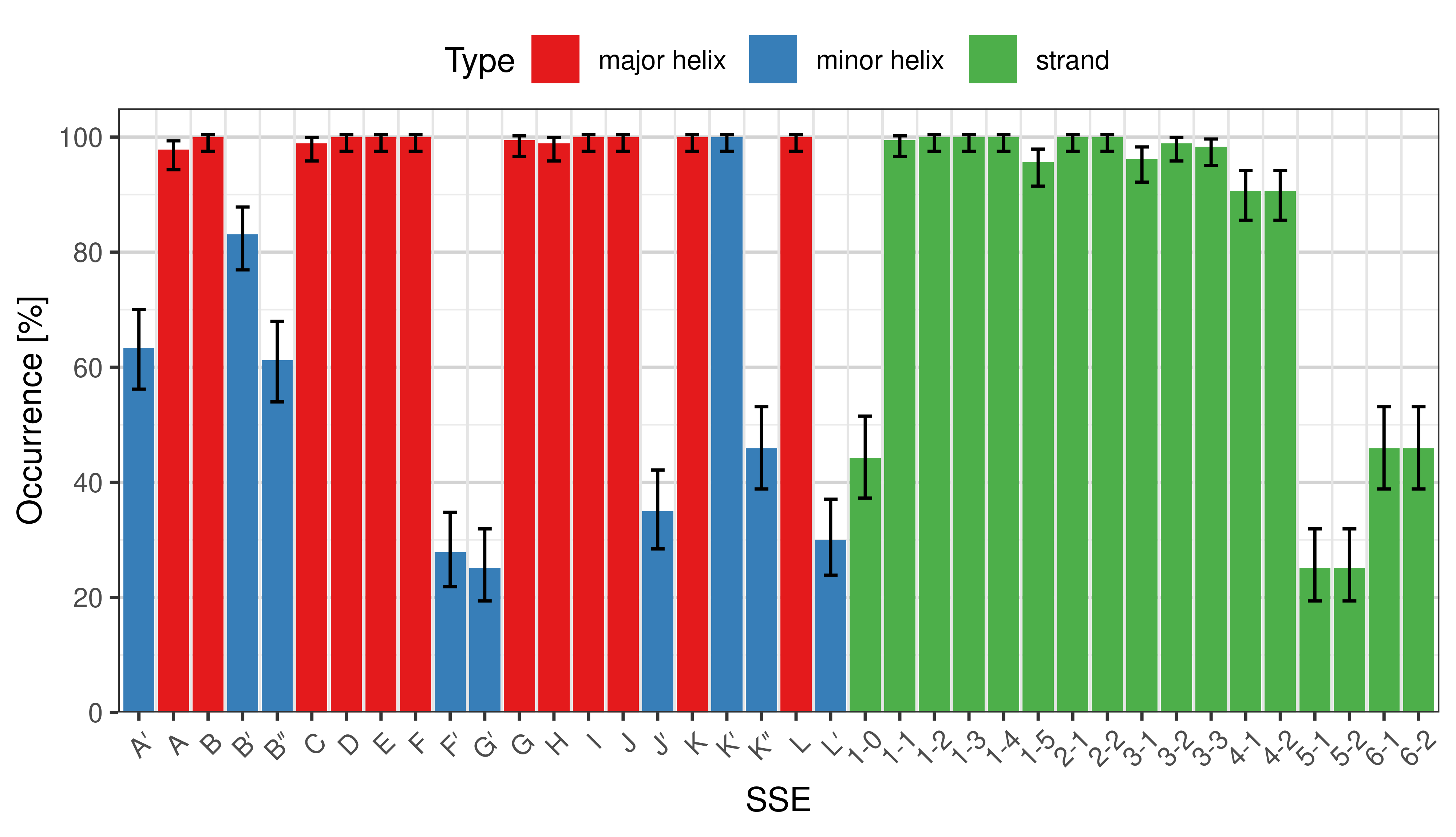 |
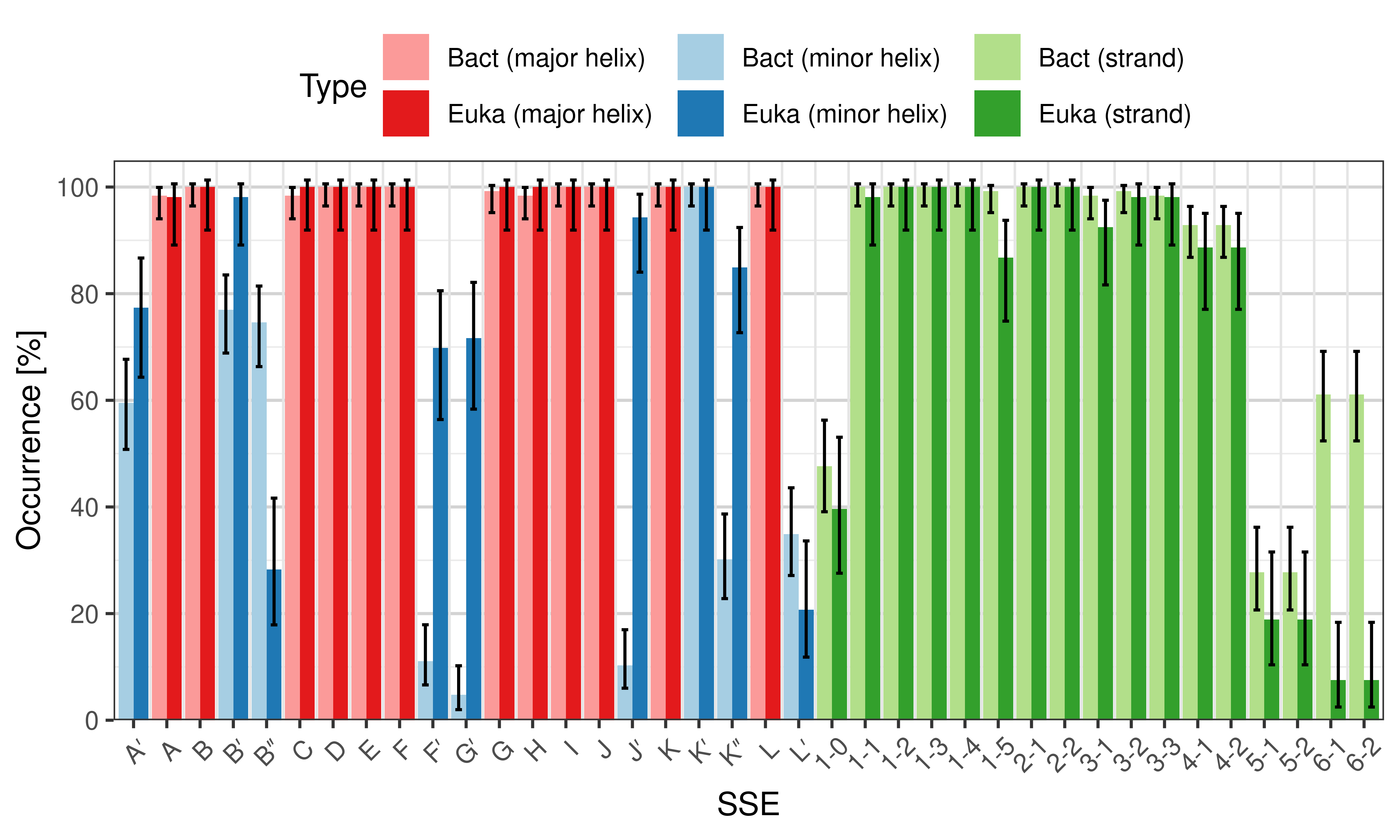 |
Length of SSEs
The length of an SSE is measured as the number of residues. The following violin plots show the distribution of length for each SSE.
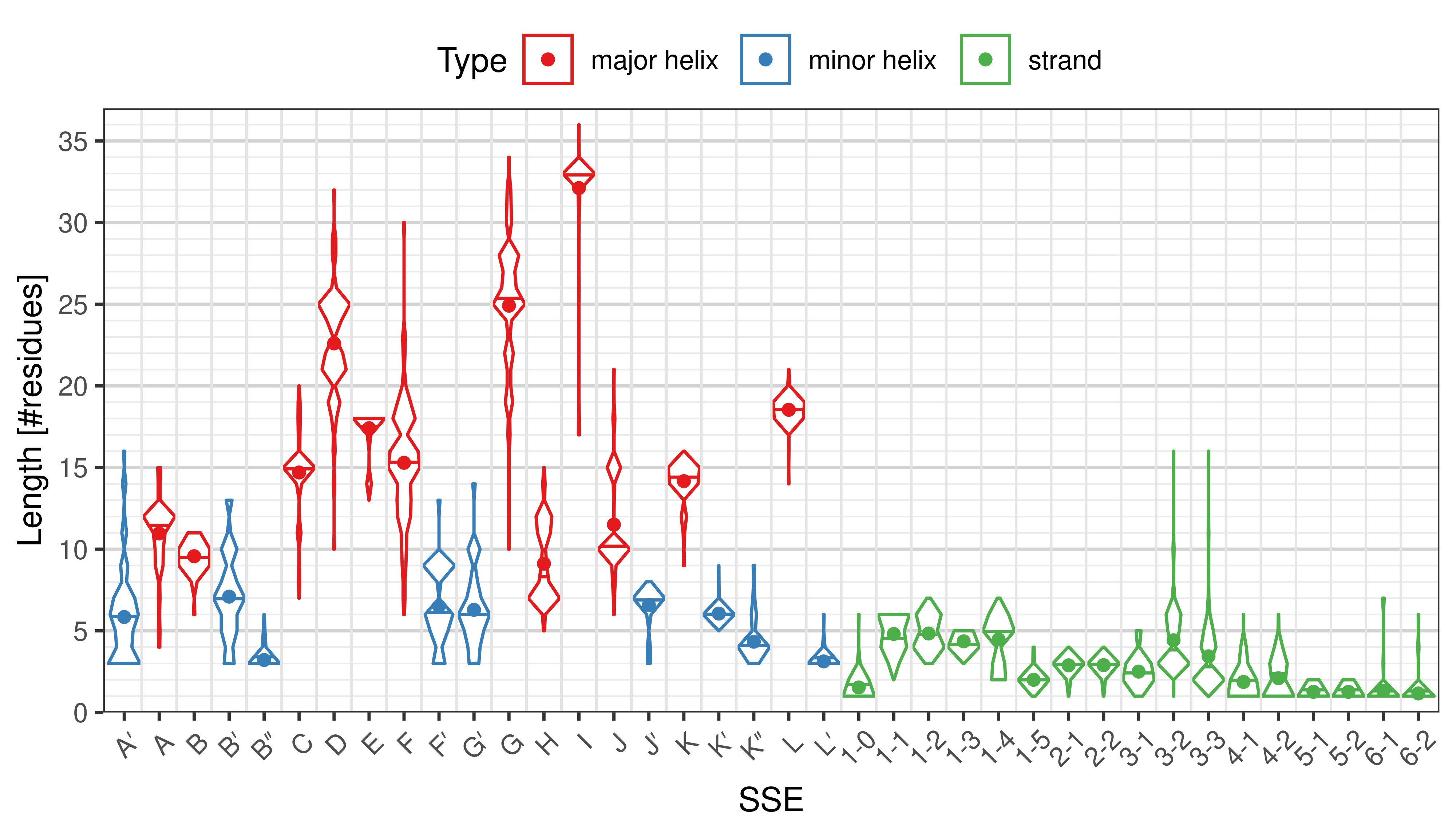 |
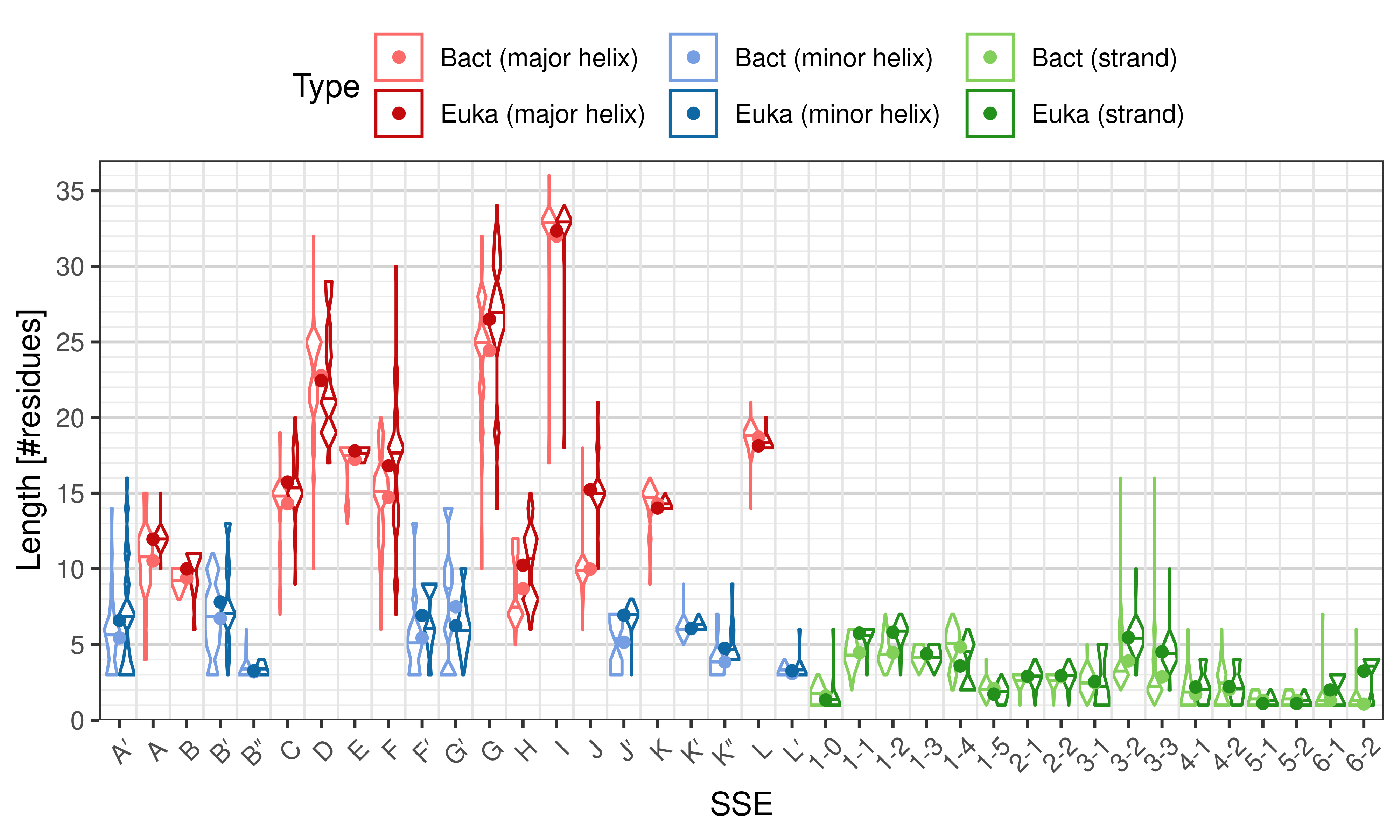 |
TODO: figure length + length-Bact-Euka (+box plots?)
Sequence of SSEs
The amino acid sequences for each SSE can be aligned and used to produce a sequence logo. Where the sequence conservation is sufficient, we can establish a generic numbering scheme: the most conserved residue in helix X serves as its reference residue and is numbered as @X.50. The remaining residues in the helix are numbered accordingly.
TODO: logos