NEEMP:Reports: Difference between revisions
Appearance
No edit summary |
No edit summary |
||
| Line 9: | Line 9: | ||
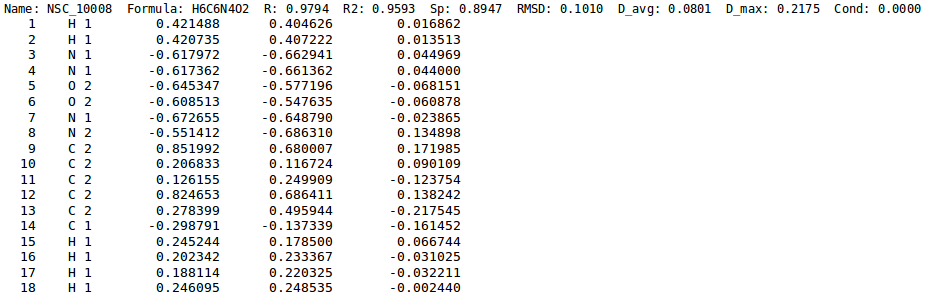
[[File:chg_stats.png | thumb | 900px | center | '''''Figure 5:''''' Close-up from charge statistics file. Along with statistics for each molecule, ''ab-initio'' charges (3rd column), ''EEM'' charges (4th column) and their difference (5th column) are also printed out.]]. | [[File:chg_stats.png | thumb | 900px | center | '''''Figure 5:''''' Close-up from charge statistics file. Along with statistics for each molecule, ''ab-initio'' charges (3rd column), ''EEM'' charges (4th column) and their difference (5th column) are also printed out.]]. | ||
''''' | Once the script has been executed, a bunch of output files are generated in the ''chg-stats-out-file'' directory. In particular: | ||
* '''''csv''''' files containing per-atom charge information and the values for several performance evaluating metrics for each atomic type and molecule | |||
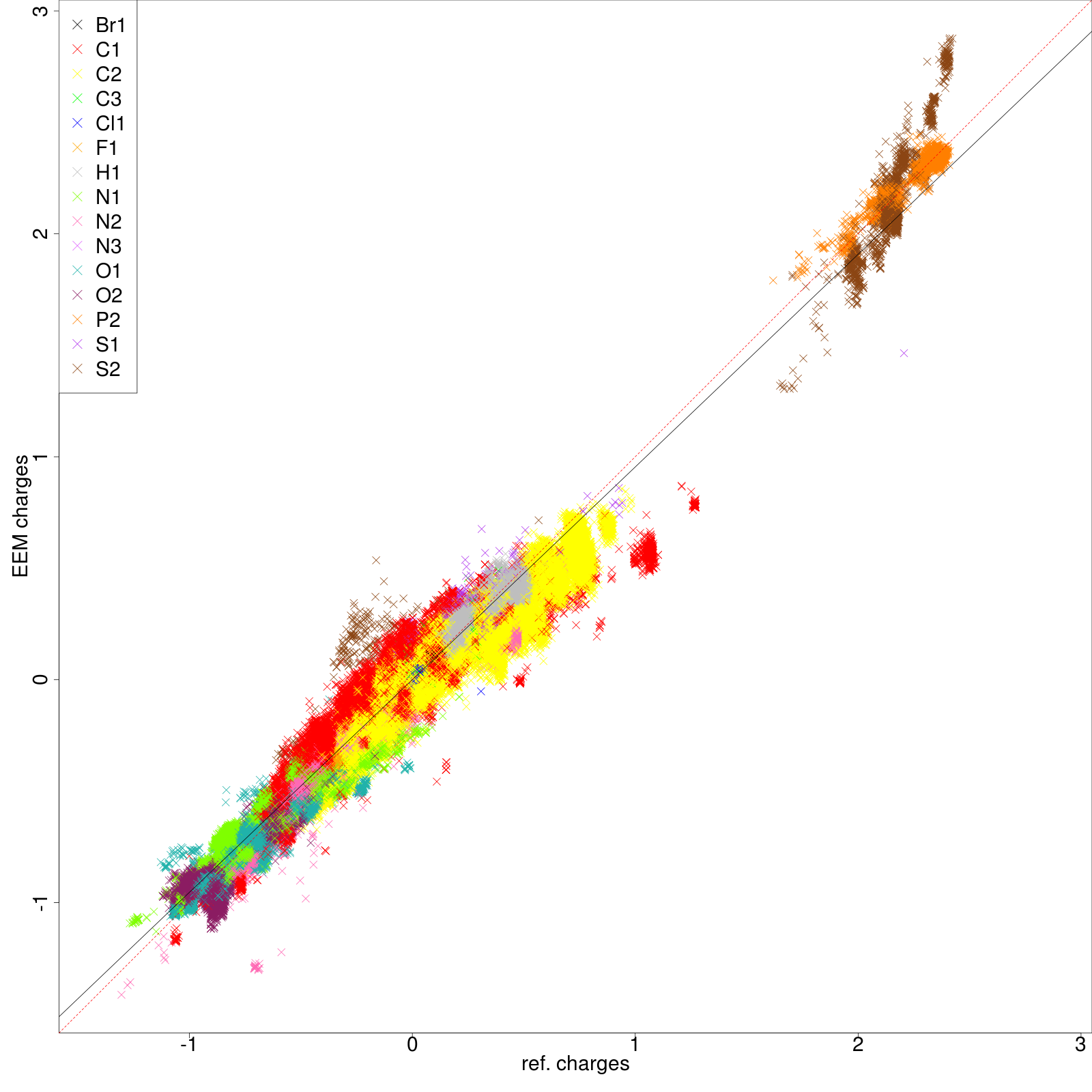
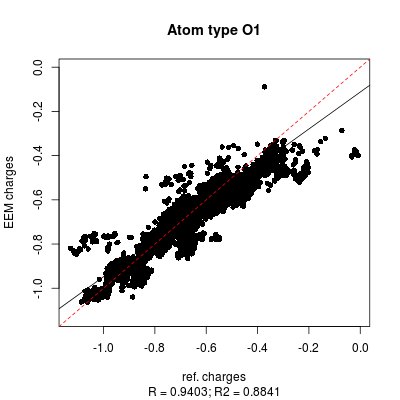
* '''''png''''' files displaying the charge correlation graphs for each atomic type and for the whole set (as shown in '''''figure 6''''' and '''''figure 7''''' respectively) | |||
* '''''html''''' file gathering together all the previous information in an interactive and more easily readable report page (see '''''figure 8''''' and '''''figure 9''''' or click [http://www.example.com here] for a more comprehensive view) | |||
{| class="wikitable" border="1" style="margin: 1em 1em 1em 1em;" width="650px" | {| class="wikitable" border="1" style="margin: 1em 1em 1em 1em;" width="650px" | ||
Revision as of 10:47, 29 June 2016
Along with NEEMP we provide an handy python script to generate charge correlation graphs and quality assay reports, namely nut-report.py. To run the above-mentioned script, python and R packages must be installed in the system.
Figure 5 displays the required input file in which for each atom the difference between the reference QM charge and the EEM charge is calculated. Such a file can be obtained running NEEMP in quality validation mode with the option --chg-stats-out (see example or quality validation for details).
SCRIPT BASIC USAGE: ./nut-report.py chg-stats-out-file
.
Once the script has been executed, a bunch of output files are generated in the chg-stats-out-file directory. In particular:
- csv files containing per-atom charge information and the values for several performance evaluating metrics for each atomic type and molecule
- png files displaying the charge correlation graphs for each atomic type and for the whole set (as shown in figure 6 and figure 7 respectively)
- html file gathering together all the previous information in an interactive and more easily readable report page (see figure 8 and figure 9 or click here for a more comprehensive view)