NEEMP:Reports: Difference between revisions
Appearance
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
All the graphs in '''NEEMP''''s article showing the correlation between reference charges and EEM charges, have been generated employing two python scripts, ''nut-report.py'' and ''nut.plot.py''. The required input file must contain the charge statistics as shown in '''''figure''''' and can be obtained running '''NEEMP''' in ''calculation'' or ''quality validation'' mode with the option <code>--chg-stats-out</code> (see [[NEEMP:Examples#Example 4 - Quality validation | here]] for details). | All the graphs in '''NEEMP''''s article showing the correlation between reference charges and EEM charges, have been generated employing two python scripts, ''nut-report.py'' and ''nut.plot.py''. The required input file must contain the charge statistics as shown in '''''figure 6''''' and can be obtained running '''NEEMP''' in ''calculation'' or ''quality validation'' mode with the option <code>--chg-stats-out</code> (see [[NEEMP:Examples#Example 4 - Quality validation | here]] for details). | ||
<br style="clear:both" /> | <br style="clear:both" /> | ||
[[File:chg_stats.png | thumb | 900px | center | '''''Figure | [[File:chg_stats.png | thumb | 900px | center | '''''Figure 6:''''' Close-up from charge statistics file. Along with statistics for each molecule, ''ab-initio'' charges (3rd column), ''EEM'' charges (4th column) and their difference (5th column) are also printed out.]]. | ||
To generate the graphs the scripts must be called in the simple following way: | To generate the graphs the scripts must be called in the simple following way: | ||
| Line 24: | Line 24: | ||
* ''./nut-plot.py set01-data.csv colorfulbig-zoomed 5'' | * ''./nut-plot.py set01-data.csv colorfulbig-zoomed 5'' | ||
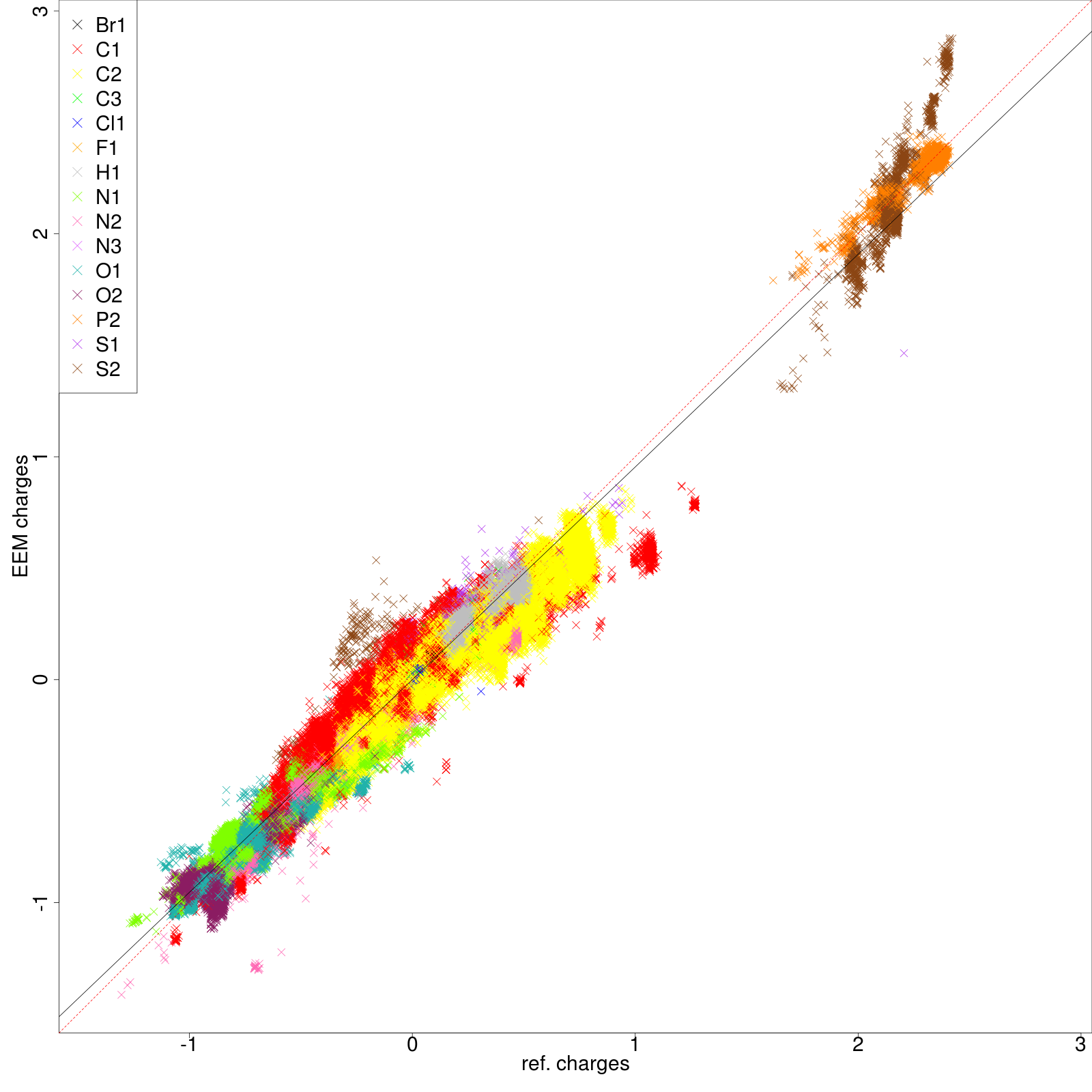
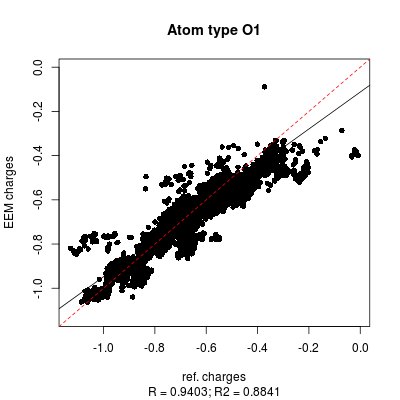
'''''Figure''''' and ''''' ''''' demonstrate as the correlation graphs include both all atom and individual atomic types dependency. | '''''Figure 7''''' and '''''8''''' demonstrate as the correlation graphs include both all atom and individual atomic types dependency. | ||
{| class="wikitable" border="1" style="margin: 1em 1em 1em 1em;" width="650px" | {| class="wikitable" border="1" style="margin: 1em 1em 1em 1em;" width="650px" | ||
| Line 31: | Line 31: | ||
| [[File:set3_DE_RMSD_B3LYP_6311G_NPA_cross_ideal_all-O1.png| 520px]] | | [[File:set3_DE_RMSD_B3LYP_6311G_NPA_cross_ideal_all-O1.png| 520px]] | ||
|- | |- | ||
|'''''Figure :''''' All atom correlation graph between ''QM charges'' (x axis) and ''EEM charges'' (y axis). | |'''''Figure 7:''''' All atom correlation graph between ''QM charges'' (x axis) and ''EEM charges'' (y axis). | ||
| '''''Figure :''''' Correlation graph between ''QM charges'' (x axis) and ''EEM charges'' (y axis) for ''O1'' atomic type (oxygen with just single bonds). | | '''''Figure 8:''''' Correlation graph between ''QM charges'' (x axis) and ''EEM charges'' (y axis) for ''O1'' atomic type (oxygen with just single bonds). | ||
|} | |} | ||
Revision as of 17:00, 23 June 2016
All the graphs in NEEMP's article showing the correlation between reference charges and EEM charges, have been generated employing two python scripts, nut-report.py and nut.plot.py. The required input file must contain the charge statistics as shown in figure 6 and can be obtained running NEEMP in calculation or quality validation mode with the option --chg-stats-out (see here for details).
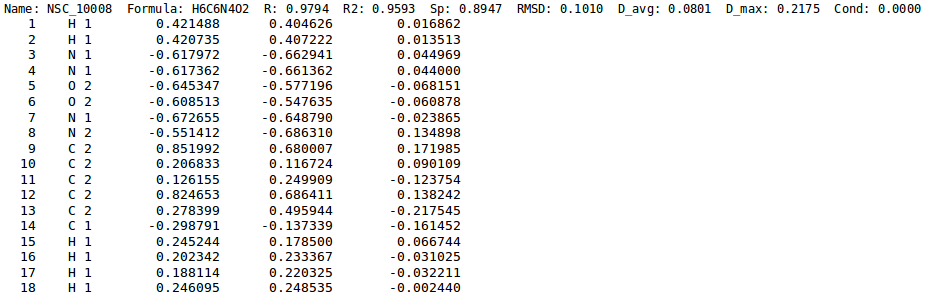
.
To generate the graphs the scripts must be called in the simple following way:
First:
- ./nut-report.py charge-stat-file
Then:
- ./nut-plot.py set01-data.csv colorful 5
- ./nut-plot.py set01-data.csv blackwhite 5
- ./nut-plot.py set01-data.csv colorfulbig 5
- ./nut-plot.py set01-data.csv colorful-zoomed 5
- ./nut-plot.py set01-data.csv blackwhite-zoomed 5
- ./nut-plot.py set01-data.csv colorfulbig-zoomed 5
Figure 7 and 8 demonstrate as the correlation graphs include both all atom and individual atomic types dependency.