NEEMP:Reports: Difference between revisions
Appearance
No edit summary |
No edit summary |
||
| Line 28: | Line 28: | ||
{| class="wikitable" border="1" style="margin: 1em 1em 1em 1em;" width="650px" | {| class="wikitable" border="1" style="margin: 1em 1em 1em 1em;" width="650px" | ||
|- | |- | ||
| [[File: | | [[File:set3_DE_RMSD_B3LYP_6311G_NPA_cross_ideal_all-summary.pgn| 620px]] | ||
| [[File: | | [[File:set3_DE_RMSD_B3LYP_6311G_NPA_cross_ideal_all-O1.png| 620px]] | ||
|- | |- | ||
|colspan=2 |'''''Figure 2:''''' Detailed view of the '''LR''' parametrization settings for two distinct '''NEEMP''' executions differing in the best-performance selection metrics (''R<sup>2</sup>'' in left side image and ''RMSD'' in the right side image). | |colspan=2 |'''''Figure 2:''''' Detailed view of the '''LR''' parametrization settings for two distinct '''NEEMP''' executions differing in the best-performance selection metrics (''R<sup>2</sup>'' in left side image and ''RMSD'' in the right side image). | ||
|} | |} | ||
Revision as of 16:46, 23 June 2016
All the graphs in NEEMP's article showing the correlation between reference charges and EEM charges, have been generated employing two python scripts, nut-report.py and nut.plot.py. The required input file must contain the charge statistics as shown in figure and can be obtained running NEEMP in calculation or quality validation mode with the option --chg-stats-out (see here for details).
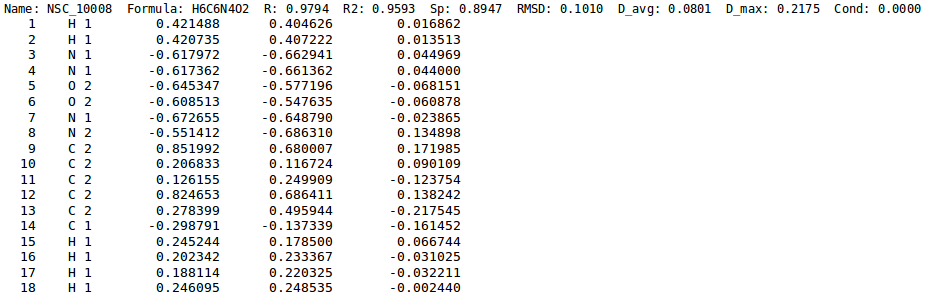
.
To generate the graphs the scripts must be called in the simple following way:
First:
- ./nut-report.py charge-stat-file
Then:
- ./nut-plot.py set01-data.csv colorful 5
- ./nut-plot.py set01-data.csv blackwhite 5
- ./nut-plot.py set01-data.csv colorfulbig 5
- ./nut-plot.py set01-data.csv colorful-zoomed 5
- ./nut-plot.py set01-data.csv blackwhite-zoomed 5
- ./nut-plot.py set01-data.csv colorfulbig-zoomed 5
Figure and demonstrate as the correlation graphs include both all atom and individual atomic types dependency.
| File:Set3 DE RMSD B3LYP 6311G NPA cross ideal all-summary.pgn | 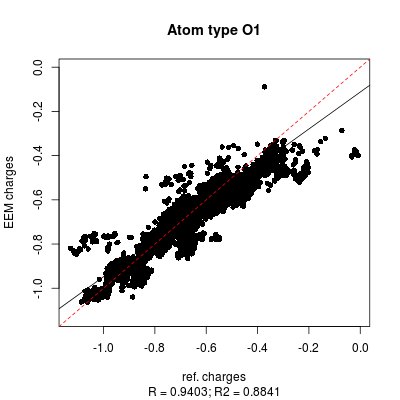
|
| Figure 2: Detailed view of the LR parametrization settings for two distinct NEEMP executions differing in the best-performance selection metrics (R2 in left side image and RMSD in the right side image). | |