NEEMP:Modes: Difference between revisions
| Line 11: | Line 11: | ||
<code>params</code> : calculate EEM parameters (due to its complexity and relevance this mode is treated on a separate [[NEEMP:parametrization | section]]) | <code>params</code> : calculate EEM parameters (due to its complexity and relevance this mode is treated on a separate [[NEEMP:parametrization | section]]) | ||
=Info ( | =Info (-m info)= | ||
[[File:info_mode.png | thumb | 400px | right | '''''Figure 1:''''' Information on training set ''examples/set01.sdf''. '''NB:''' Information about the execution time and the percentage of loaded molecules is issued as well.]] | [[File:info_mode.png | thumb | 400px | right | '''''Figure 1:''''' Information on training set ''examples/set01.sdf''. '''NB:''' Information about the execution time and the percentage of loaded molecules is issued as well.]] | ||
Revision as of 15:25, 28 May 2016
NEEMP features several functionalities, each of those can be selected by the option --mode or -m combined with one of the following keywords:
info : display information about the training set
cover : display how many molecules are covered by the given parameters set
charges : calculate EEM charges
cross : perform cross-validation
params : calculate EEM parameters (due to its complexity and relevance this mode is treated on a separate section)
Info (-m info)
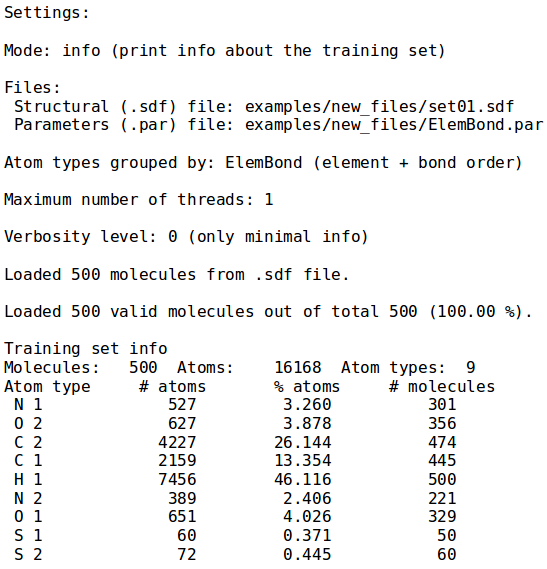
The first mode shows information about the training set provided as argument to the --sdf-file option. Specifically, number of molecules, the total number of atoms and number of atoms for each type are printed; the latter being defined by default as the chemical element and the maximum bond order. To override this behaviour in all NEEMP's modes and group the atoms only by chemical element, use the option --atom-types-by Element. Figure 1 displays the output for set01 in the examples directory, with default grouping, specified by ElemBond keyword.