PatternQuery:How to build a query: Difference between revisions
No edit summary |
|||
| Line 1: | Line 1: | ||
<div class="toclimit-4"> | <div class="toclimit-4"> | ||
In order to tune up your queries before executing them on the whole database or just to | In order to tune up your queries before executing them on the whole database or just to work with the language interactively, feel free to use the [http://webchem.ncbr.muni.cz/Platform/MotiveQuery/Index '''Explorer'''] application, where you can upload a PDB molecule of choice, or load a random sample from the PDB database based on selected properties. | ||
You can either try one of our ready-to-use examples or try to make up one of your own. | You can either try one of our ready-to-use examples or try to make up one of your own. | ||
==How to think about queries== | ==How to think about queries== | ||
When building queries, you have to decompose the problem | When building queries, you have to decompose the problem into smaller chunks, think of a query for each of the pieces and then wisely bind them together. Let us have a goal: | ||
'''Get all the residues which are as much as 5Å away from any histidine residue and check if the given fragment contains at least 2 negatively charged amino acids.''' | '''Get all the residues which are as much as 5Å away from any histidine residue and check if the given fragment contains at least 2 negatively charged amino acids.''' | ||
From scratch you can't probably think out a solution right now, but when you decompose the problem into separate individual sub problems, it is not so difficult after all. (Whenever you are unsure about the meaning of the queries, consult the [[MotiveQuery:Language Reference | language reference]]). For given problem, the decomposition is as follows: | From scratch you can't probably think out a solution right now, but when you decompose the problem into separate individual sub problems, it is not so difficult after all. (Whenever you are unsure about the meaning of the queries, consult the [[MotiveQuery:Language Reference | language reference]]). For the given problem, the decomposition is as follows: | ||
#Select histidine residues | #Select histidine residues | ||
| Line 37: | Line 37: | ||
==Ready-to-use examples== | ==Ready-to-use examples== | ||
Now when we are aware of how to think about queries, you are ready to browse a plenty of different examples listed below. The text is separated | Now when we are aware of how to think about queries, you are ready to browse a plenty of different examples listed below. The text is separated into three plus one part which differs by data type queries are operated (Atoms, Residues and Fragments). The first two categories deal with only basic ''Atom'' or ''Residue'' selections. Outputs of these two types of queries are Atoms and Residues respectively. Last category is 'Fragments', which contains a number of advanced queries, these queries demonstrate versatility and a power of '''MQ'''. These queries operate on all results provided by both Atom and Residue queries. On the top of that, you can browse several use biologically relevant [[MotiveQuery:Use Cases | use cases]]. | ||
| Line 182: | Line 182: | ||
====RegularMotifs()==== | ====RegularMotifs()==== | ||
* '''[PDB id: 1het]''' | * '''[PDB id: 1het]''' Sequence motifs is extracted from the primary sequence based on the input regular expression. Please note, that MQ does not check presence of a gap gap in the chain e.g. if HIS 28 is followed by the residue 30 ALA query ‘HA’ returns positive match for such example. | ||
* | * In case the information about posttranslational modifications is present in ''MODRES'' or ''_pdbx_struct_mod_residue'' fields, modified residues are treated as standard amino acids. Therefore, a letter ''P'' stands for a proline residue so as all its modifications, e.g. '''HYP''' (hydroxyproline). | ||
* <code> RegularMotifs("HH")</code> – Finds two consecutive histidine residues or their modifications. | * <code> RegularMotifs("HH")</code> – Finds two consecutive histidine residues or their modifications. | ||
* <code> RegularMotifs("G.{1,2}G")</code> – Finds two glycine residues separated by one or two other residues. | * <code> RegularMotifs("G.{1,2}G")</code> – Finds two glycine residues separated by one or two other residues. | ||
* <code> RegularMotifs("G.{1,2}G").Filter(lambda m: m.IsConnected())</code> – Finds two glycine residues separated by one or two residues and verifies that all of them are bonded. | * <code> RegularMotifs("G.{1,2}G").Filter(lambda m: m.IsConnected())</code> – Finds two glycine residues separated by one or two residues and verifies that all of them are bonded. | ||
* <code> RegularMotifs(".P.").Filter(lambda l: l.Count(NotAminoAcids()) == 0)</code> - Finds | * <code> RegularMotifs(".P.").Filter(lambda l: l.Count(NotAminoAcids()) == 0)</code> - Finds 3 consecutive residues, where the middle one is proline and verifies that neither of them is outside the ''standard 20''. | ||
* ''Type: RegularMotifs(regex: Value) -> Fragments'' | * ''Type: RegularMotifs(regex: Value) -> Fragments'' | ||
====Cluster()==== | ====Cluster()==== | ||
* Clusters | * Clusters identifies results to a single fragment based on their distance [Å]. On contrary to the <code>Near()</code> query <code>Cluster()</code> does not provide a count check. See example below. | ||
* <code>Cluster(5, Residues("Ala"))</code> – Returns all the alanine residues which are at most 5A distant to each other. | * <code>Cluster(5, Residues("Ala"))</code> – Returns all the alanine residues which are at most 5A distant to each other. | ||
* <code>Cluster(3, RingAtoms(Atoms("N"), Rings(2 * ["C"] + ["N"] + ["C"] + ["N"])))</code> – Returns all the nitrogen atoms from particular rings to a single fragment in case the atoms are at most 3A distant to each other. | * <code>Cluster(3, RingAtoms(Atoms("N"), Rings(2 * ["C"] + ["N"] + ["C"] + ["N"])))</code> – Returns all the nitrogen atoms from particular rings to a single fragment in case the atoms are at most 3A distant to each other. | ||
* <code>Cluster(2 , Atoms("C"), Atoms("C"))</code> – Returns all the carbon atoms which are pairwise closer than | * <code>Cluster(2 , Atoms("C"), Atoms("C"))</code> – Returns all the carbon atoms which are pairwise closer than 2Å. In case any other carbon atom would be inside this 2Å sphere, it is also included in the result. Therefore, if you insist on exactly 2 carbon atoms to be returned, use <code>Near()</code> query instead. | ||
* ''Type: Cluster(r: Number, fragments: FragmentSeq) -> Fragments'' | * ''Type: Cluster(r: Number, fragments: FragmentSeq) -> Fragments'' | ||
| Line 207: | Line 207: | ||
===Topology function=== | ===Topology function=== | ||
====AmbientAtoms()==== | ====AmbientAtoms()==== | ||
* '''[PDB id: 1hho]''' Returns all the atoms, which are within | * '''[PDB id: 1hho]''' Returns all the atoms, which are within nÅ from the geometrical center of a given fragment. | ||
* <code>Atoms("Fe").AmbientAtoms(4)</code> – returns all the atoms, which are closer than | * <code>Atoms("Fe").AmbientAtoms(4)</code> – returns all the atoms, which are closer than 4Å from the center of mass of iron atom. | ||
* ''Type: AmbientAtoms(fragment: FragmentSeq, r: Number) -> Fragments'' | * ''Type: AmbientAtoms(fragment: FragmentSeq, r: Number) -> Fragments'' | ||
| Line 214: | Line 214: | ||
====AmbientResidues()==== | ====AmbientResidues()==== | ||
* '''[PDB id: 1hho]''' Returns all the residues, which are within | * '''[PDB id: 1hho]''' Returns all the residues, which are within nÅ from the geometrical center of a given fragment. | ||
* <code>Residues("HEM").AmbientResidues(4)</code> – returns all the residues, which are closer than | * <code>Residues("HEM").AmbientResidues(4)</code> – returns all the residues, which are closer than 4Å from the center of mass of HEM residues. | ||
* ''Type: AmbientResidues(fragment: FragmentSeq, r: Number) -> Fragments' | * ''Type: AmbientResidues(fragment: FragmentSeq, r: Number) -> Fragments' | ||
====Near()==== | ====Near()==== | ||
* '''[PDB id: 1hho]''' Clusters all the specified fragments, which are pairwise closer | * '''[PDB id: 1hho]''' Clusters all the specified fragments, which are pairwise closer [Å] than a specified argument. Additionally, it checks if the fragment contains exactly specified fragments. On the contrary to the Cluster() query this does the ‘count check’. | ||
* <code>Near(0, Rings(6*['C']), Rings(4*['C'] + ['N']))</code> – Returns fragments containing only purine. In this example the purine part of tryptophan side chain will be returned. | * <code>Near(0, Rings(6*['C']), Rings(4*['C'] + ['N']))</code> – Returns fragments containing only purine. In this example the purine part of tryptophan side chain will be returned. | ||
* <code>Near(2 , Atoms("C"), Atoms("C"), Atoms("C"), Atoms("C"))</code> – Returns fragments which contain exactly 4 carbon atoms within a sphere of | * <code>Near(2 , Atoms("C"), Atoms("C"), Atoms("C"), Atoms("C"))</code> – Returns fragments which contain exactly 4 carbon atoms within a sphere of 2Å. | ||
* ''Type: Near(r: Number, fragments: FragmentSeq+) -> Fragments'' | * ''Type: Near(r: Number, fragments: FragmentSeq+) -> Fragments'' | ||
| Line 227: | Line 227: | ||
===Filtering=== | ===Filtering=== | ||
====Filter()==== | ====Filter()==== | ||
* '''[PDB id: 1hho]''' | * '''[PDB id: 1hho]''' Filtering is used for removing unwanted fragments from the result. A condition given as a function argument is evaluated for each fragment from input fragment sequence referred to as ‘m’ in the following text. Only results satisfying the condition are returned, others are filtered out. Technically, it uses lambda abstraction for filtering a collection of input fragments. The usage is the same as in the Python programming language, and therefore, your previous Python experiences are beneficial. | ||
* <code>Residues().Filter(lambda m: m.IsConnectedTo(Atoms("Fe"))) </code> – returns a set of fragments from all residues which are covalently bonded to the iron atom (excluded the HEM residue). | * <code>Residues().Filter(lambda m: m.IsConnectedTo(Atoms("Fe"))) </code> – returns a set of fragments from all residues which are covalently bonded to the iron atom (excluded the HEM residue). | ||
* <code> Residues().Filter(lambda m: m.Count(Atoms("O")) == 2)</code> returns all the residues containing exactly two oxygen atoms. | * <code> Residues().Filter(lambda m: m.Count(Atoms("O")) == 2)</code> returns all the residues containing exactly two oxygen atoms. | ||
| Line 237: | Line 237: | ||
* <code>Residues("CYS").ConnectedResidues(1).Filter(lambda m: m.Count(Residues("VAL")) == 2)</code> – Returns a cysteine residue which is surrounded from both sides by valine residues. | * <code>Residues("CYS").ConnectedResidues(1).Filter(lambda m: m.Count(Residues("VAL")) == 2)</code> – Returns a cysteine residue which is surrounded from both sides by valine residues. | ||
* ''Type: Count(where: Fragment, what: FragmentSeq) -> Integer''. | * ''Type: Count(where: Fragment, what: FragmentSeq) -> Integer''. | ||
Revision as of 15:19, 20 April 2015
In order to tune up your queries before executing them on the whole database or just to work with the language interactively, feel free to use the Explorer application, where you can upload a PDB molecule of choice, or load a random sample from the PDB database based on selected properties.
You can either try one of our ready-to-use examples or try to make up one of your own.
How to think about queries
When building queries, you have to decompose the problem into smaller chunks, think of a query for each of the pieces and then wisely bind them together. Let us have a goal:
Get all the residues which are as much as 5Å away from any histidine residue and check if the given fragment contains at least 2 negatively charged amino acids.
From scratch you can't probably think out a solution right now, but when you decompose the problem into separate individual sub problems, it is not so difficult after all. (Whenever you are unsure about the meaning of the queries, consult the language reference). For the given problem, the decomposition is as follows:
- Select histidine residues
- Select residual surrounding of a fragment up to 5Å.
- Find negatively charged residues.
- Count the amino acids.
- Filter the histidine plus its surrounding matching the condition above.
Now we are ready for constructing individual queries:
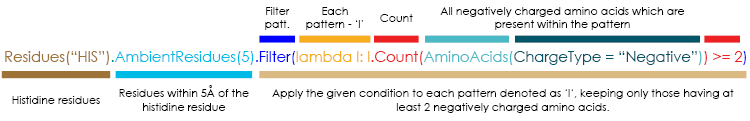
Residues("HIS")AmbientResidues(5)AminoAcids(ChargeType="Negative")Count(residues)Filter(condition)
It wasn't that difficult, was it? When we have composed all the queries, we can compose them together in order to achieve our goal as highlighted in the query bellow and in the illustrative info-graphics:
Residues("HIS").
AmbientResidues(5).
Filter(lambda l: l.Count(AminoAcids(ChargeType = "Negative")) >= 2)
Ready-to-use examples
Now when we are aware of how to think about queries, you are ready to browse a plenty of different examples listed below. The text is separated into three plus one part which differs by data type queries are operated (Atoms, Residues and Fragments). The first two categories deal with only basic Atom or Residue selections. Outputs of these two types of queries are Atoms and Residues respectively. Last category is 'Fragments', which contains a number of advanced queries, these queries demonstrate versatility and a power of MQ. These queries operate on all results provided by both Atom and Residue queries. On the top of that, you can browse several use biologically relevant use cases.
Structure of the text
- [PDB id] Here you find an example PDB id where you can try out the query with MotiveQuery Explorer and a rough description of the query function.
- This is followed by examples of this query with with the explanation such as
Residues("HEM"). Copy this query to the command text box in MotiveQuery Explorer application and immediately see the results. - Type of data query operate on and the expected returned value.
- e.g. Type: Atoms(symbols: String*) -> Atoms. query
Atoms()take 0..n strings, representing elements, in parenthesis ("C", "N", etc.) and based on the input returns a list of individual atoms.
Queries
Atoms
Basic queries
Atoms()
- [PDB id: 2hhb] Returns a sequence of individual atoms based on element type provided in the argument. More elements can be specified, if separated by a comma. In case no argument is provided a list of all the atoms is returned.
Atoms("Fe")- Returns all iron atoms in the given structure.Atoms("Fe", "N")- Returns all iron and nitrogen atoms in the given structure.- Type: Atoms(symbols: String*) -> Atoms.
AtomNames()
- [PDB id: 2hhb] Returns a sequence of atoms with defined name or names.
AtomNames("CA")– Returns all CA atoms.AtomNames("CA", "N")–Returns atoms with names CA (C? carbon) or N (terminal part of amino acids).- Type: AtomNames(names: String+) -> Atoms.
AtomIds()
- [PDB id: 2hhb] Returns a sequence of atoms with given id or ids
AtomIds(1)– Returns atom with id=1 from the given structures.AtomIds(1,2,5)- Returns atoms with id=1, 2 and 5 from the given structures.- Type: AtomIds(ids: Integer+) -> Atoms.
AtomIdRange()
- [PDB id: 2hhb] Returns a sequence of atoms with ids from a given range (inclusive specified indices).
AtomIdRange(1, 10)– returns 10 atoms with IDs from the interval <1, 10>, as specified in the input file.- Type: AtomIdRange(minId: Integer, maxId: ?Integer) -> Atoms
NotAtomNames()
- [PDB id: 2hhb] Returns a sequence of atoms which are not defined by an argument.
NotAtomNames("C", "N", "CA", "O")– returns all the atoms with names other than C, CA, N and O. i.e. only the side chain atoms of the protein .- Type: NotAtomNames(names: String+) -> Atom.
NotAtomIds()
- [PDB id: 2hhb] Returns a sequence of atoms which does not have a defined id
NotAtomIds(1)- Returns atom with id other but 1 from the given structures.NotAtomIds(1,2,5)- Returns atoms with id other but 1,2 and 5 from the given structures.- Type: NotAtomIds(ids: Integer+) -> Atoms.
NotAtoms()
- [PDB id: 2hhb] Returns all the atoms not specified in the argument. More elements can be specified, if separated by a comma.
Atoms("Fe")– returns all the atoms of the structure, but iron.Atoms("Fe", "N")returns all the atoms of the structure, but iron and nitrogen.- Type: NotAtoms(symbols: String+) -> Atoms.
RingAtoms()
- [PDB id: 2hhb] Returns specified atoms found on detected rings .
RingAtoms(Atoms("N"), Rings(2 * ["C"] + ["N"] + ["C"] + ["N"]))– Returns all the nitrogen atoms on the histidine side chain.- Type: RingAtoms(atom: Atoms, ring: ?Rings) -> Atoms.
Residues
Basic queries
Residues()
- [PDB id: 2hhb] Returns a sequence of individual residues specified by a function argument. More residues can be specified, if separated by a comma.
Residues("HEM")– Returns a list of HEM residues.Residues ("HEM", "ALA")– Returns a set of HEM and ALA residues.- Type: Residues(names: Value*) -> Residues.
NotResidues()
- [PDB id: 2hhb] Returns a sequence of residues which are not defined by the argument.
NotResidues("HEM")– returns a set of residues, which does not have a HEM in their name.- Type: NotResidues(names: Value+) -> Residues.
ResidueIds()
- [PDB id: 2hhb] Returns a sequence of specified residues in case the structure contains them. Each residue is represented by its PDB ID and chain such as “A 8”.
ResidueIds ("14 A", "15 A")– Returns 14th and 15th residue of chain A.- Type: ResidueIds(ids: String+) -> Residues.
ResidueIdRange()
- [PDB id: 2hhb] Returns a set of residues on a given chain from the lower to the upper index. In case a residue is not provided in the structure, it is skipped.
ResidueIdRange("A", 50, 100)– returns a set of residues on chain A from the ID 50 to 100.- Type: ResidueIdRange(chain: String, min: Integer, max: Integer) -> Residues.
NotAminoAcids()
- [PDB id: 2hhb] Returns a sequence of residues that are not among the 20 standard amino acids. Allowed values for an optional parameter NoWaters: True, False.
NotAminoAcids()– Returns all the nonstandard residues incorporated in the protein structure with the exception of HOH and WAT residues, which stands for solvent.NotAminoAcids(NoWaters=False)– Returns all the nonstandard residues incorporated in the protein structure inclusive solvent (HOH and WAT residues).- Type: NotAminoAcids() -> Residues.
AminoAcids()
- [PDB id: 2hhb] Returns a sequence of residues that are among the 20 standard amino acids.
- Allowed values:
Positive, Negative, Aromatic, Polar, NonPolar AminoAcids()– Returns all standard amino acids.AminoAcids(ChargeType="Polar")– Returns all polar amino acids based on the type of their side chain.- Type: AminoAcids() -> Residues.
HetResidues()
- [PDB id: 2hhb] Returns a sequence of heteroatom residues as specified in input PDB files, excluding residues.
HetResidues()– A set of hetatom residues.- Type: HetResidues() -> Residues
Fragments
Structure specification
MQ can select residues or their parts based on their name or Id, however, the true power of MQ lies in its ability to utilize the chemical nature of the input structures and select fragments purely based on elements and the connectivity among them.
Rings()
- [PDB id: 3d12] Returns all the rings in the protein structure specified by a user. Any structural ring can be identified by concatenating individual elements the string is composed from.
Rings(2 * ["C"] + ["N"] + ["C"] + ["N"])– Returns the histidine aromatic ring.Rings(4 * ["C"] + ["O"])– Returns pentose ring.- Type: Rings(atoms: Value*) -> Ring
ToAtoms()
- [PDB id: 1hho] Converts the input fragment into a sequence of individual atoms, i.e. each fragment contains a single atom.
Residues("HEM").ToAtoms()– Returns all the atoms of HEM residues as a sequence of individual atoms.- Type: ToAtoms(fragments: Fragments) -> Fragments
ToResidues()
- [PDB id: 1hho] Converts the input fragment into a sequence of individual residues, i.e. It can either decompose a fragment with multiple residues to a sequence of fragments each containing a single residue, or if applied to a sequence of atoms merge atoms to a single fragment per residue.
Residues("HEM").AmbientResidues(2).ToResidues()– Returns the sequence of individual residues from the 2A surrounding of the HEM residue, inclusive HEM.Atoms("C").ToResidues()– Returns a sequence of fragments. Each fragment contains only carbon atoms grouped together according to their parent residue.- Type: ToResidues(fragments: FragmentSeq) -> Fragments
Union()
- [PDB id: 1hho] Merges the sequence of input fragments to a single fragment.
Residues("HEM").ConnectedResidues(1).Union()– Takes two HEM residues of the 1hho protein with covalently bonded residues (2 fragments) and merges them into a single fragment.- Type: Union(fragments: FragmentSeq) -> Fragments
RegularMotifs()
- [PDB id: 1het] Sequence motifs is extracted from the primary sequence based on the input regular expression. Please note, that MQ does not check presence of a gap gap in the chain e.g. if HIS 28 is followed by the residue 30 ALA query ‘HA’ returns positive match for such example.
- In case the information about posttranslational modifications is present in MODRES or _pdbx_struct_mod_residue fields, modified residues are treated as standard amino acids. Therefore, a letter P stands for a proline residue so as all its modifications, e.g. HYP (hydroxyproline).
RegularMotifs("HH")– Finds two consecutive histidine residues or their modifications.RegularMotifs("G.{1,2}G")– Finds two glycine residues separated by one or two other residues.RegularMotifs("G.{1,2}G").Filter(lambda m: m.IsConnected())– Finds two glycine residues separated by one or two residues and verifies that all of them are bonded.RegularMotifs(".P.").Filter(lambda l: l.Count(NotAminoAcids()) == 0)- Finds 3 consecutive residues, where the middle one is proline and verifies that neither of them is outside the standard 20.- Type: RegularMotifs(regex: Value) -> Fragments
Cluster()
- Clusters identifies results to a single fragment based on their distance [Å]. On contrary to the
Near()queryCluster()does not provide a count check. See example below. Cluster(5, Residues("Ala"))– Returns all the alanine residues which are at most 5A distant to each other.Cluster(3, RingAtoms(Atoms("N"), Rings(2 * ["C"] + ["N"] + ["C"] + ["N"])))– Returns all the nitrogen atoms from particular rings to a single fragment in case the atoms are at most 3A distant to each other.Cluster(2 , Atoms("C"), Atoms("C"))– Returns all the carbon atoms which are pairwise closer than 2Å. In case any other carbon atom would be inside this 2Å sphere, it is also included in the result. Therefore, if you insist on exactly 2 carbon atoms to be returned, useNear()query instead.- Type: Cluster(r: Number, fragments: FragmentSeq) -> Fragments
Boolean operations
Or()
- [PDB id: 2hhb] Merges up to n different fragments together.
Or(AminoAcids(ChargeType="Negative"),AminoAcids(ChargeType= "Positive"))– returns all charged amino acids both positively charged and negatively charged.- Type: Or(fragments: FragmentSeq+) -> Fragments
Topology function
AmbientAtoms()
- [PDB id: 1hho] Returns all the atoms, which are within nÅ from the geometrical center of a given fragment.
Atoms("Fe").AmbientAtoms(4)– returns all the atoms, which are closer than 4Å from the center of mass of iron atom.- Type: AmbientAtoms(fragment: FragmentSeq, r: Number) -> Fragments
AmbientResidues()
- [PDB id: 1hho] Returns all the residues, which are within nÅ from the geometrical center of a given fragment.
Residues("HEM").AmbientResidues(4)– returns all the residues, which are closer than 4Å from the center of mass of HEM residues.- Type: AmbientResidues(fragment: FragmentSeq, r: Number) -> Fragments'
Near()
- [PDB id: 1hho] Clusters all the specified fragments, which are pairwise closer [Å] than a specified argument. Additionally, it checks if the fragment contains exactly specified fragments. On the contrary to the Cluster() query this does the ‘count check’.
Near(0, Rings(6*['C']), Rings(4*['C'] + ['N']))– Returns fragments containing only purine. In this example the purine part of tryptophan side chain will be returned.Near(2 , Atoms("C"), Atoms("C"), Atoms("C"), Atoms("C"))– Returns fragments which contain exactly 4 carbon atoms within a sphere of 2Å.- Type: Near(r: Number, fragments: FragmentSeq+) -> Fragments
Filtering
Filter()
- [PDB id: 1hho] Filtering is used for removing unwanted fragments from the result. A condition given as a function argument is evaluated for each fragment from input fragment sequence referred to as ‘m’ in the following text. Only results satisfying the condition are returned, others are filtered out. Technically, it uses lambda abstraction for filtering a collection of input fragments. The usage is the same as in the Python programming language, and therefore, your previous Python experiences are beneficial.
Residues().Filter(lambda m: m.IsConnectedTo(Atoms("Fe")))– returns a set of fragments from all residues which are covalently bonded to the iron atom (excluded the HEM residue).Residues().Filter(lambda m: m.Count(Atoms("O")) == 2)returns all the residues containing exactly two oxygen atoms.- Type: Filter(fragments: FragmentSeq, filter: Fragment->Bool) -> Fragments
Count()
- [PDB id: 1hho] Usually it is convenient to utilize this function inside a filtering query.
The Count()query counts the number of occurrences of a fragment inside a different fragment. Residues("HEM").ConnectedResidues(2).Filter(lambda m: m.Count(Atoms("S")) == 1)– Returns fragments composed of a HEM residue surrounded by two layers of bonded residues in case that the whole fragment contains exactly one sulphur atom.Residues("CYS").ConnectedResidues(1).Filter(lambda m: m.Count(Residues("VAL")) == 2)– Returns a cysteine residue which is surrounded from both sides by valine residues.- Type: Count(where: Fragment, what: FragmentSeq) -> Integer.
Contains()
- [PDB id: 4hhb]
Contains()query checks if the input fragment contains a specified fragment of interest. In other words Contains() query is similar to a query, where the number of occurrences is higher than zero(Count() > 0) Residues("HEM").AmbientResidues(2).Filter(lambda m: m.Contains(Residues("HIS")))– Returns fragments, where any atom of HEM residue is at most 2A distant from the histidine.Residues().Filter(lambda m: m.Contains(Atoms("S")))– Returns all the residue which has a sulphur incorporated in their structure. For this particular example it is similar to the queryResidues("CYS", "MET").- Type: Contains(where: Fragment, what: FragmentSeq) -> Bool
Connectivity
IsConnected()
- [PDB id: 4m9e, 4m9v] Checks, whether a particular fragment is composed of a single component
Atoms("Zn").AmbientAtoms(4).Filter(lambda m: m.IsConnected())– Atoms which are at most 4 A distant from the zinc atom and are all binded together, i.e. there is no outlier.- For comparison please compare with the results of the query
Atoms("Zn").AmbientAtoms(4) - Type: IsConnected(fragment: Fragment) -> Bool.
IsConnectedTo()
- [PDB id: 1hho] Checks if the two provided fragments are connected one to another.
Residues("ALA").Filter(lambda m: m.IsConnectedTo(Residues("GLY")))– Returns all the alanine residues and directly connected glycine residues.- Type: IsConnectedTo(current: Fragment, fragments: FragmentSeq) -> Bool
IsNotConnectedTo()
- [PDB id: 1hho] Checks if the two provided fragments are NOT connected one to another.
Residues("ALA").Filter(lambda m: m.IsNotConnectedTo(Residues("GLY")))– Returns all the alanine residues which are not directly connected to the glycine residues.- Type: IsNotConnectedTo(current: Fragment, fragments: FragmentSeq) -> Bool
ConnectedAtoms()
- [PDB id: 1hho] Returns n directly bonded layer of atoms to a given fragment.
Atoms("Fe").ConnectedAtoms(1)– The iron atom and all the atoms which are covalently bonded to it over a single bond.Atoms("Fe"). ConnectedAtoms (2)– Previous selection and all the atoms which are covalently bonded to them (i.e. additional layer of bonded atom). In other words the output composes of all the atoms which are 2 bonds away from the iron atom.- Type: ConnectedAtoms(fragment: FragmentSeq, n: Integer) -> Fragments
ConnectedResidues()
- [PDB id: 1hho] Returns n directly bonded layer of residues to a given fragment.
Residues("HEM").ConnectedResidues(1)– The HEM residue and all the residues which are covalently bonded to any atom of the HEM residue.Residues("HEM").ConnectedResidues(2)– previous selection and all the residues which are covalently bonded to them (i.e. additional layer of bonded residues).- Type: ConnectedResidues(fragment: FragmentSeq, n: Integer) -> Fragments