SecStrAnnotator:Analysis: Difference between revisions
No edit summary |
|||
| Line 34: | Line 34: | ||
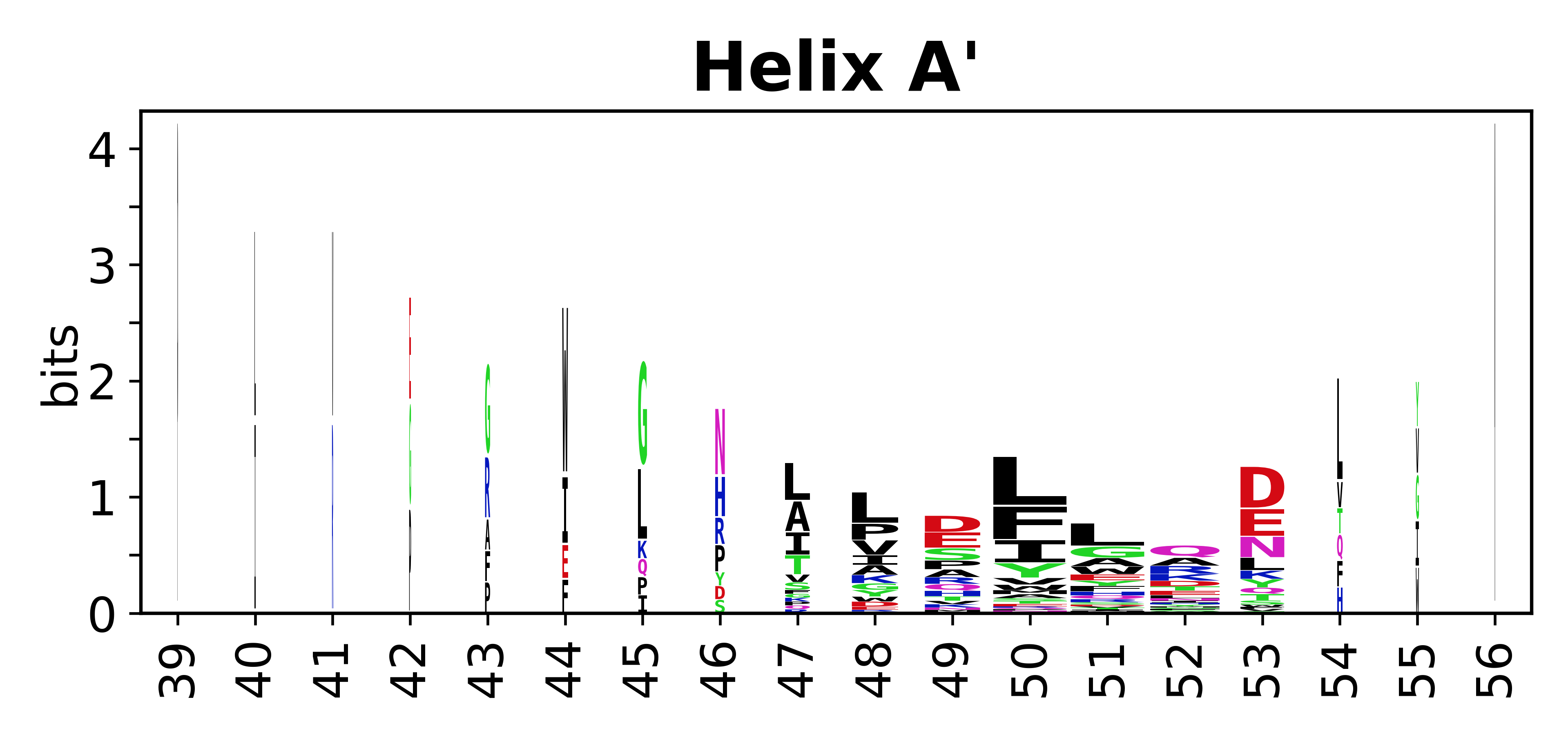
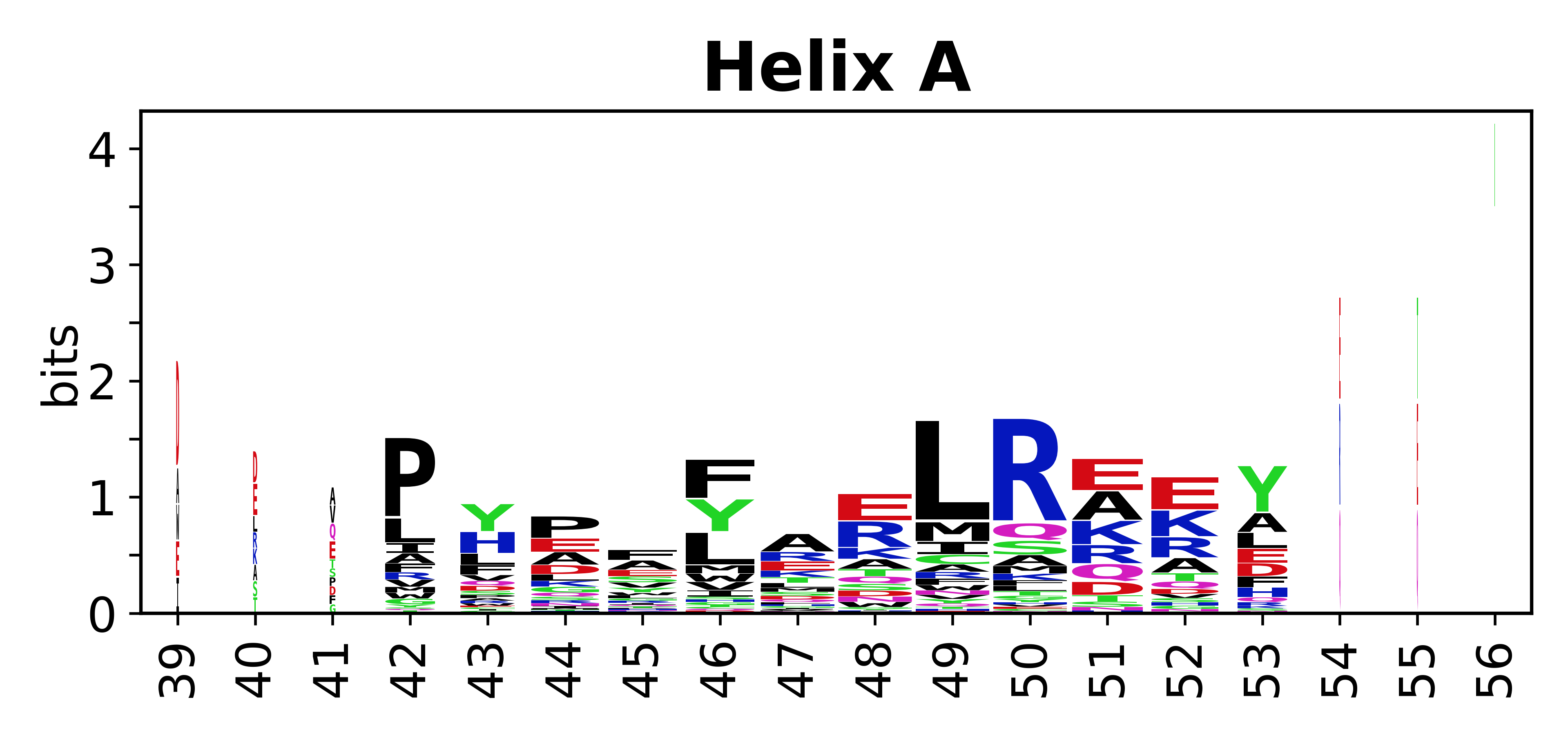
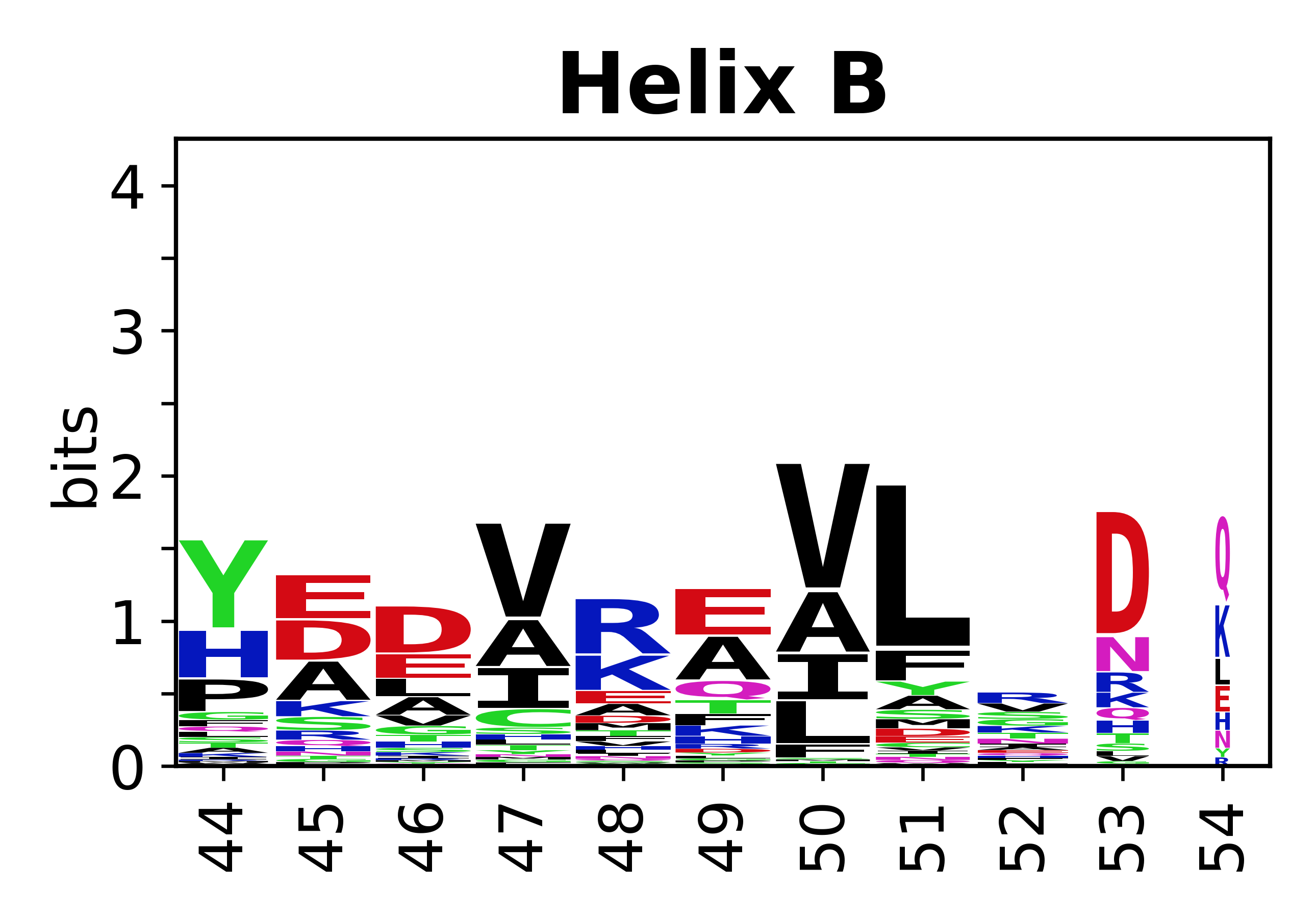
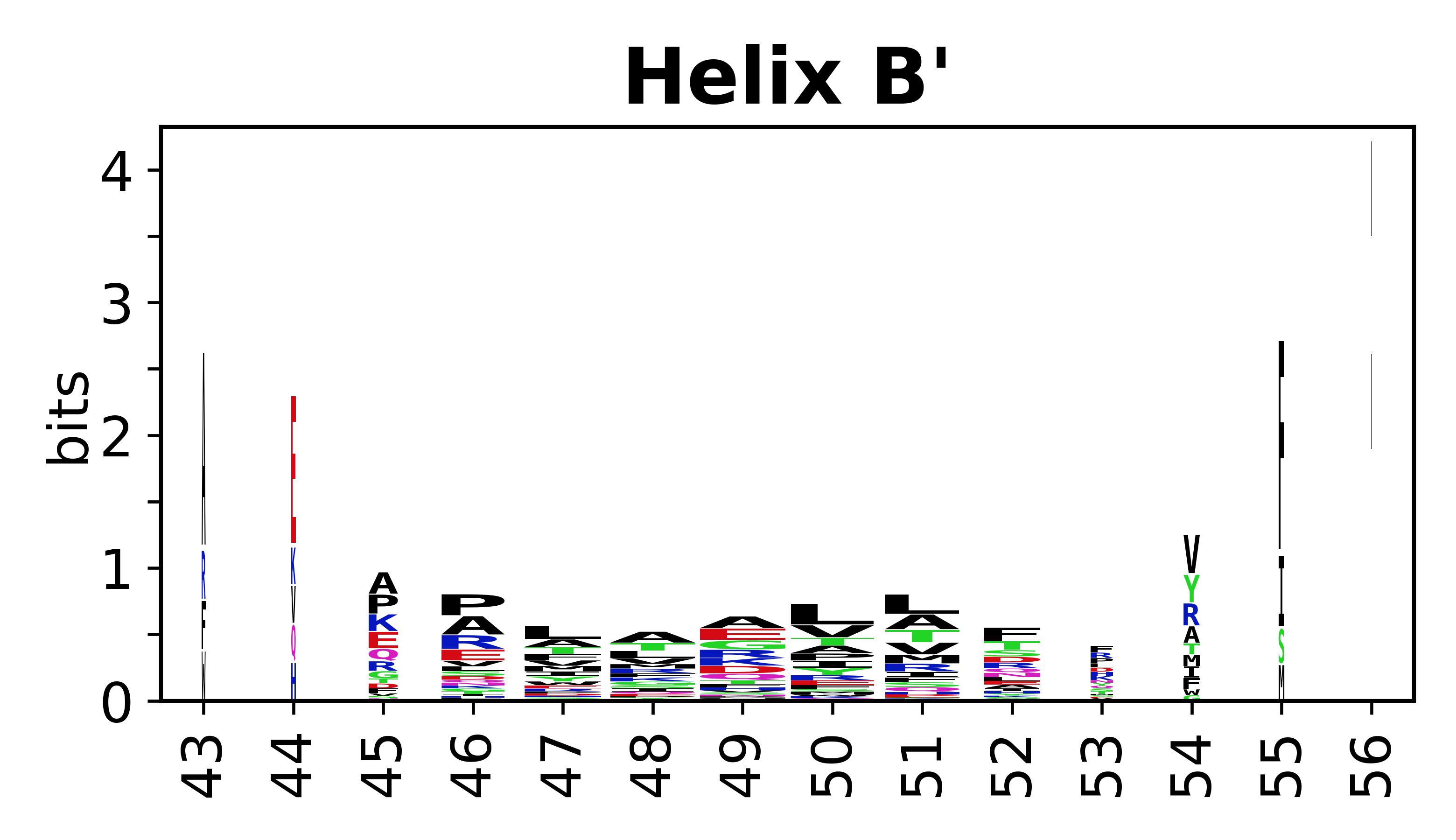
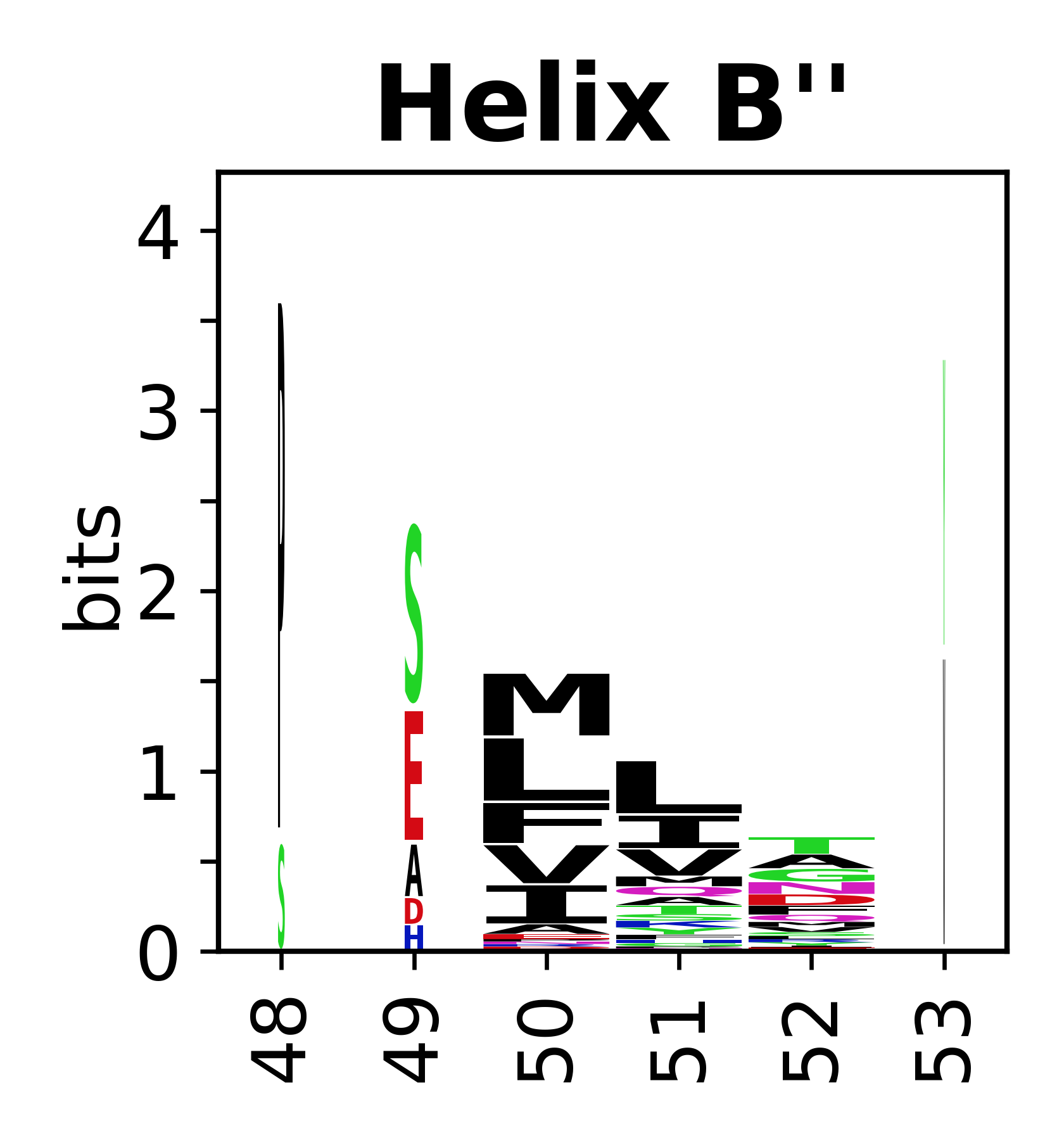
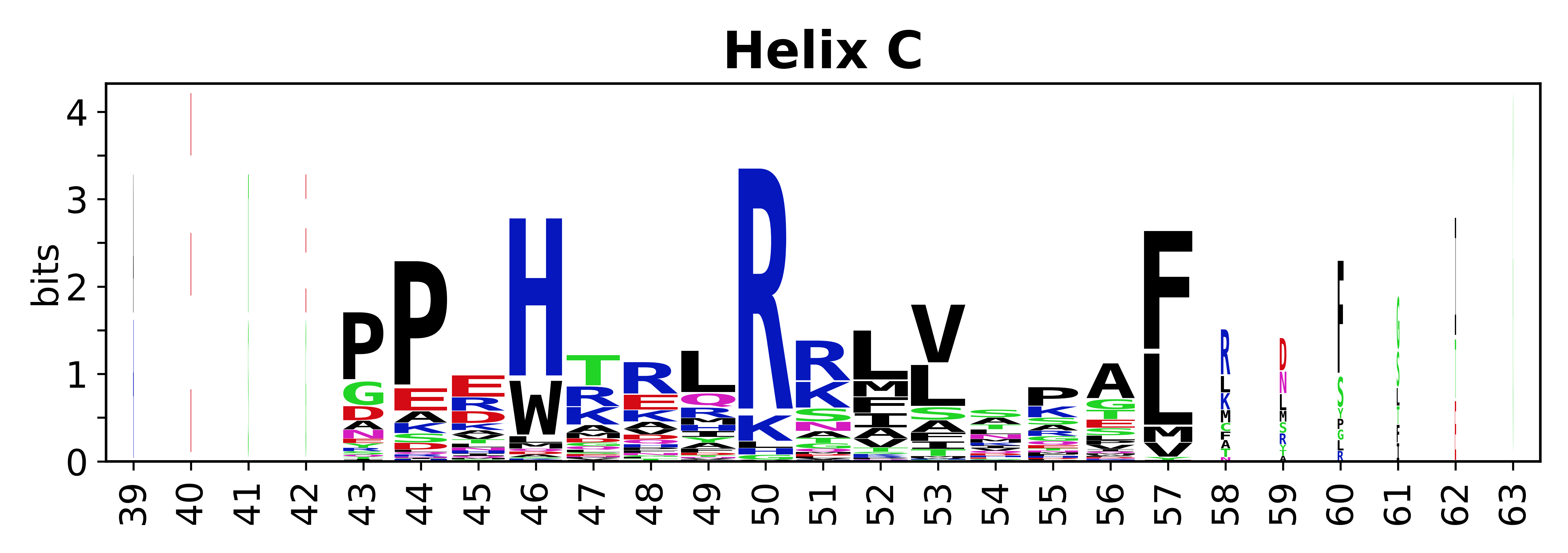
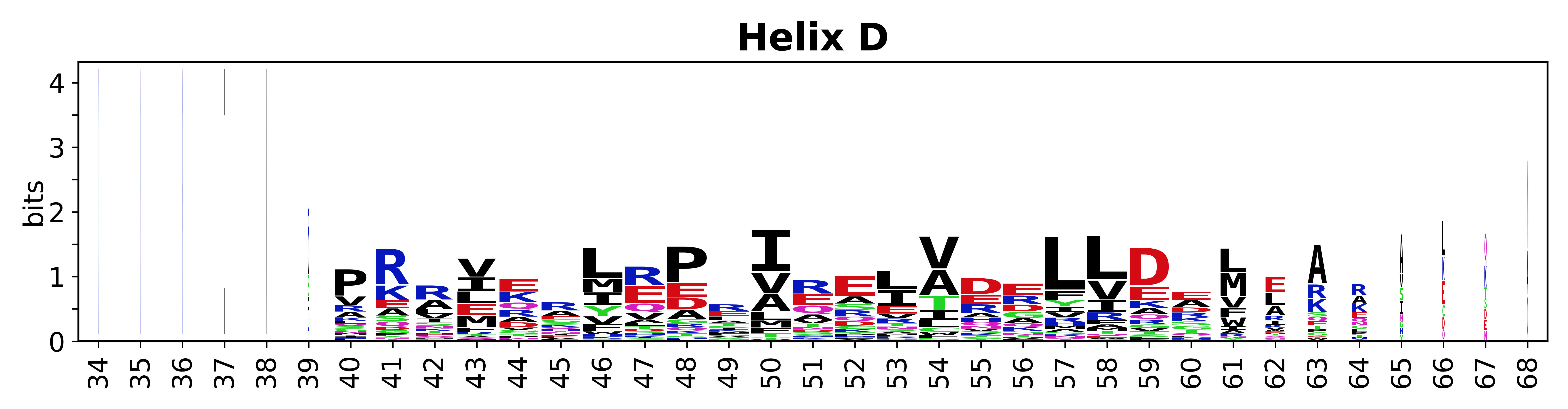
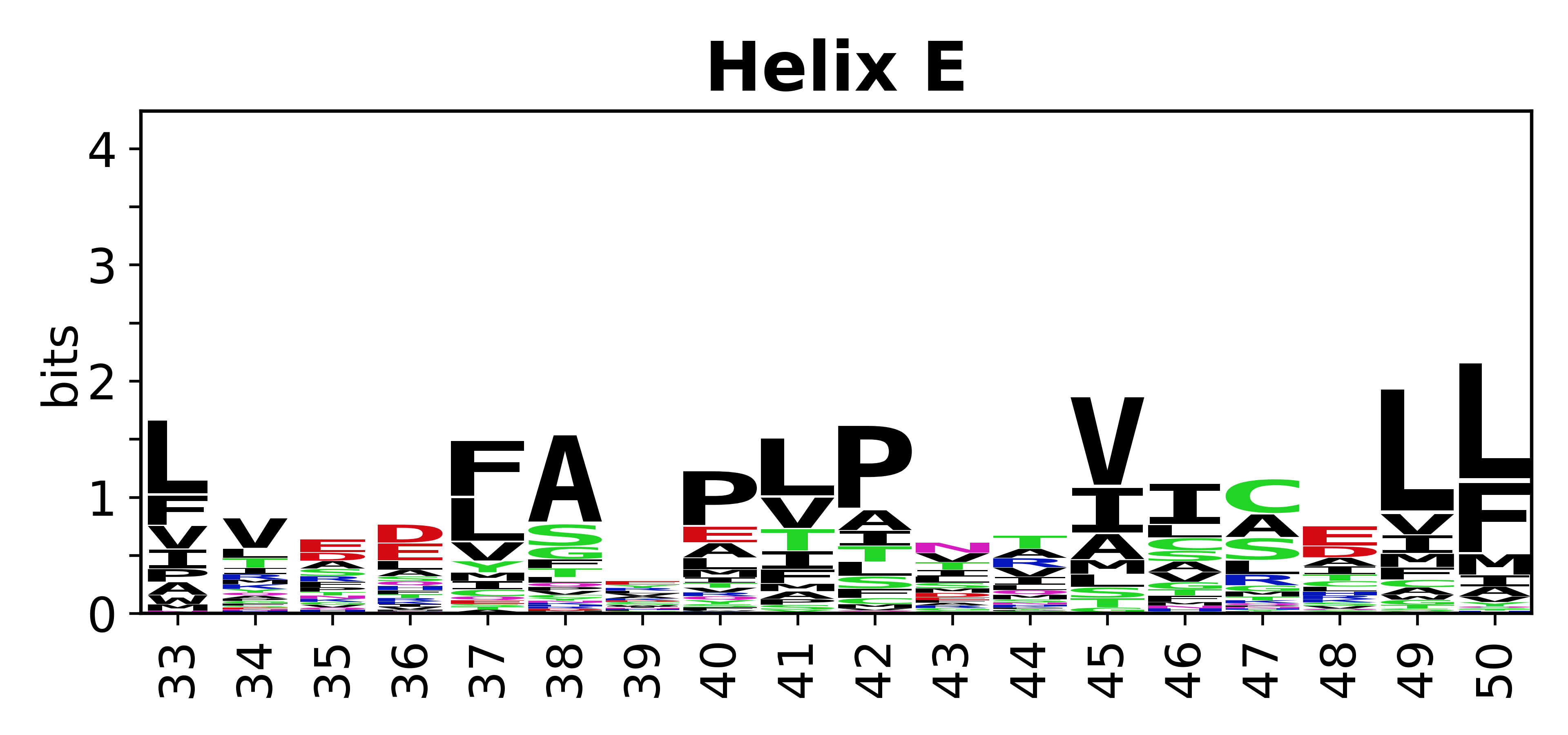
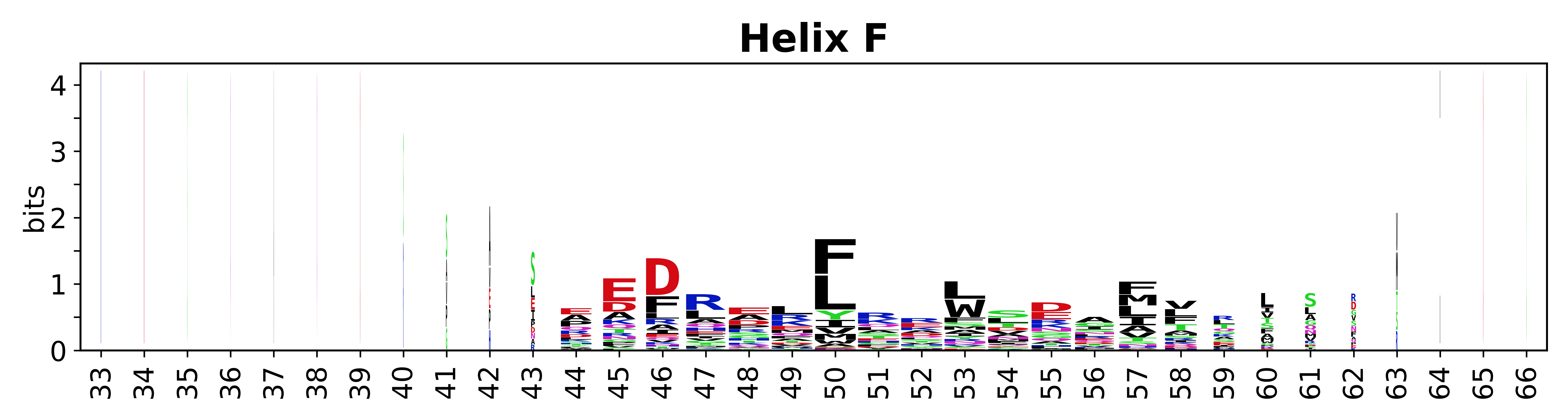
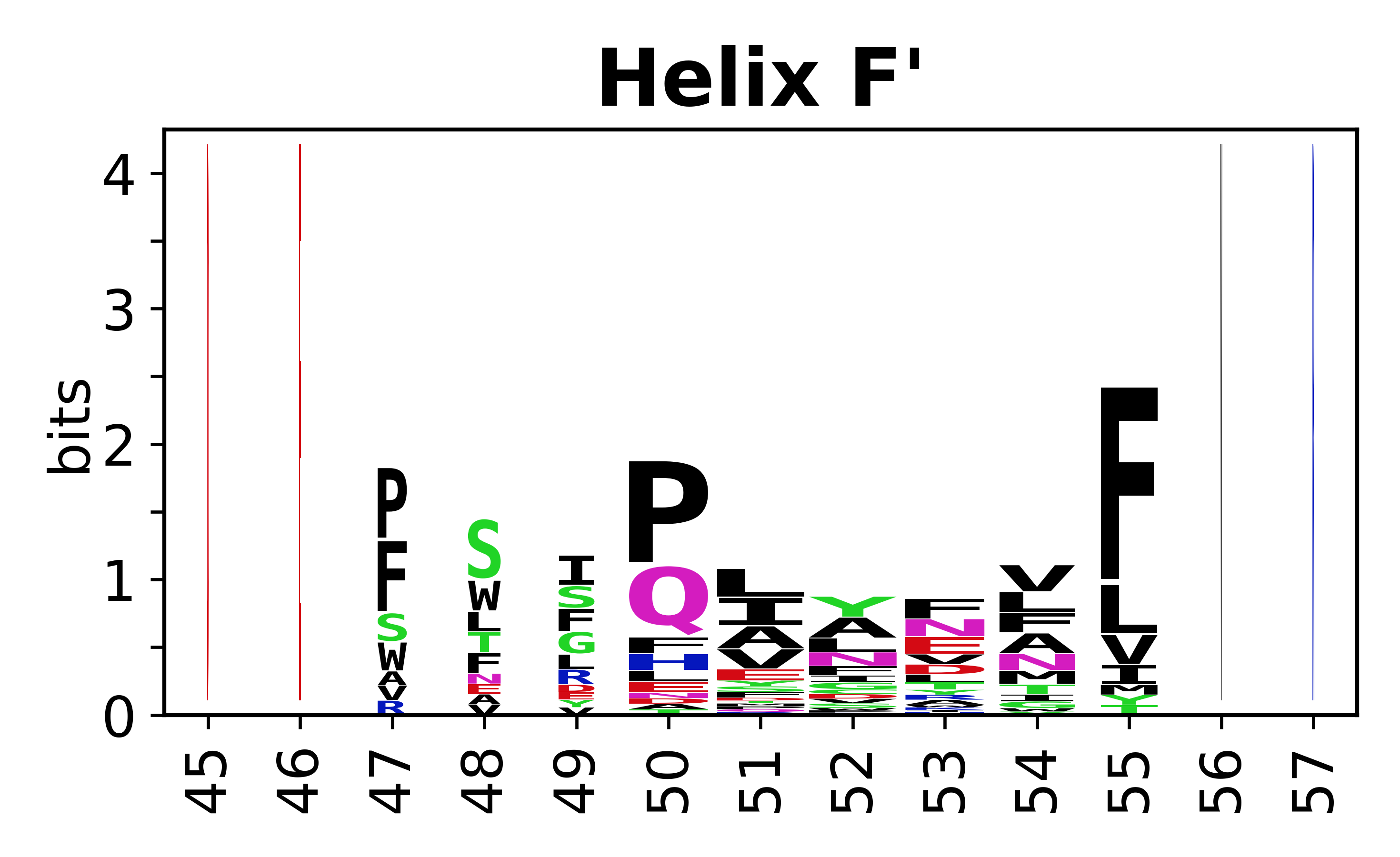
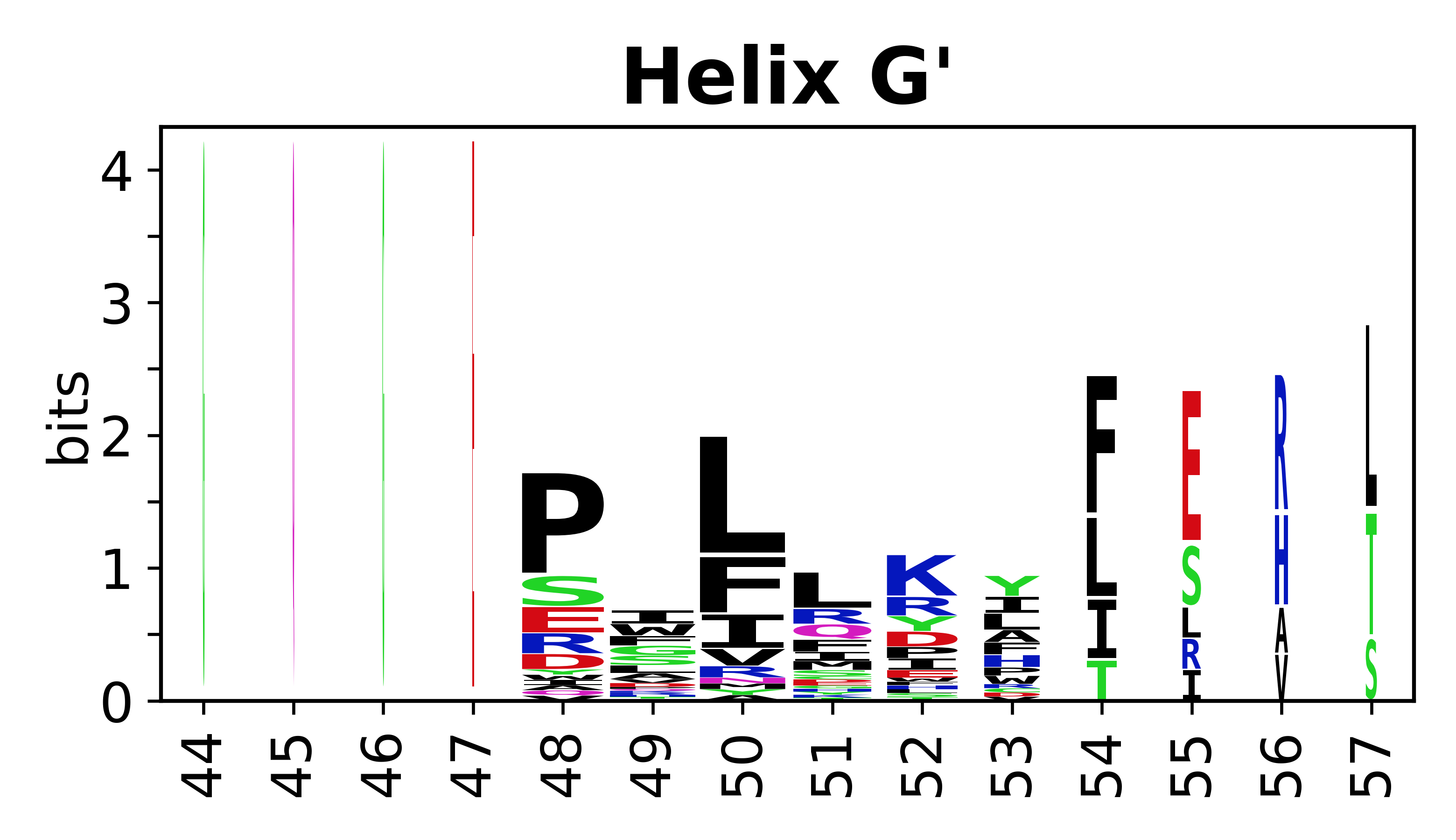
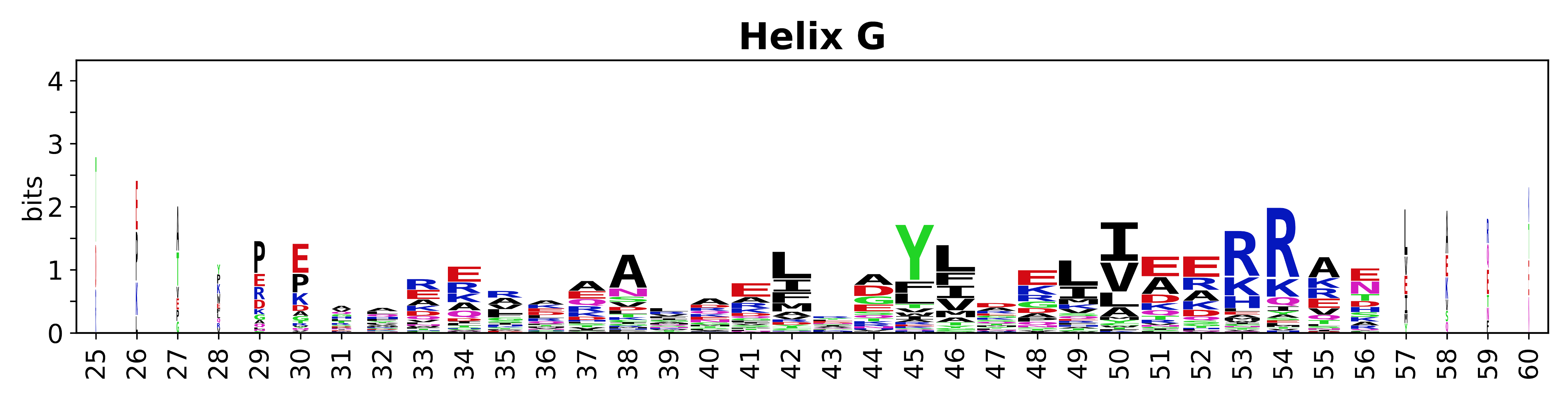
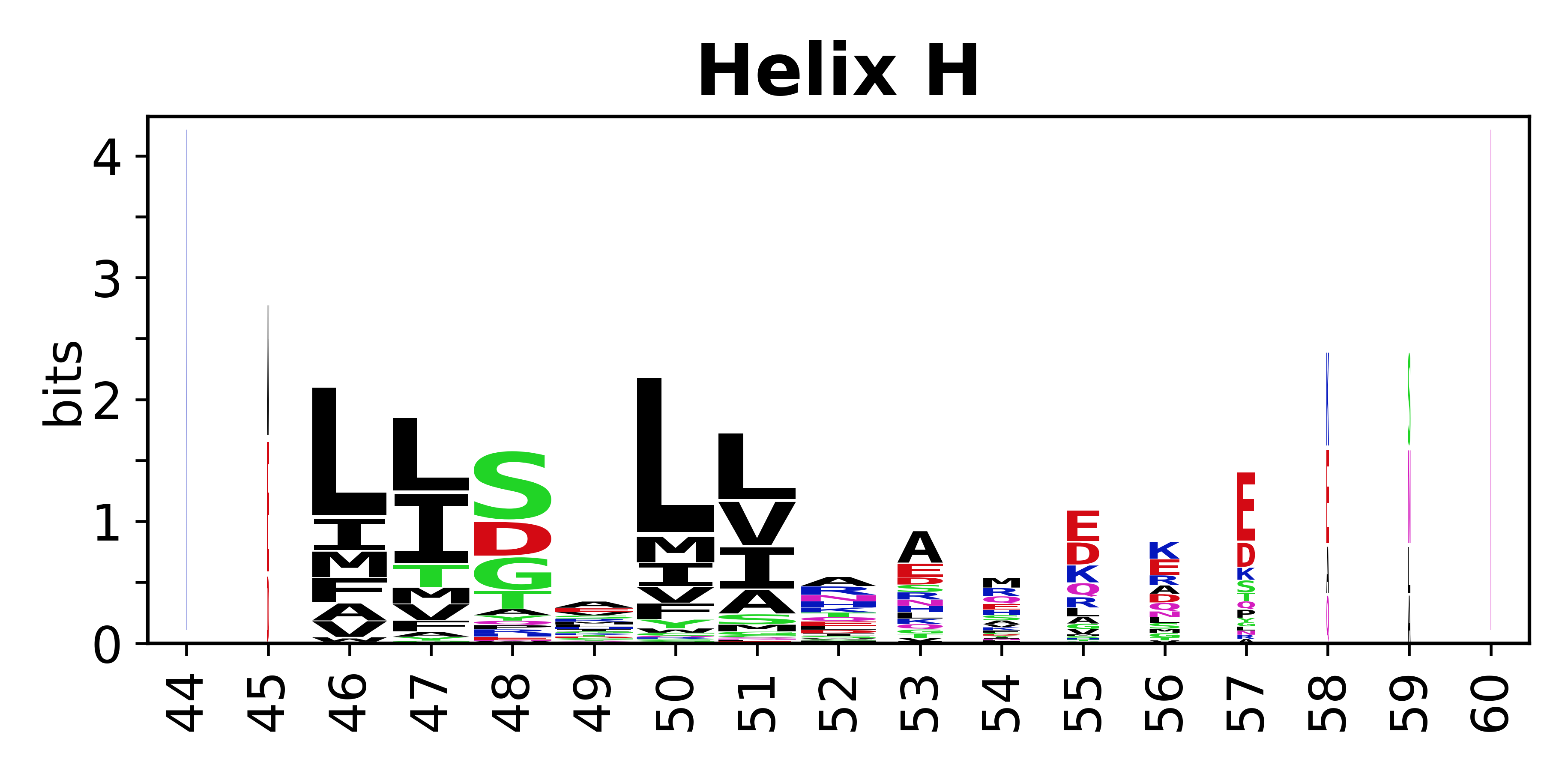
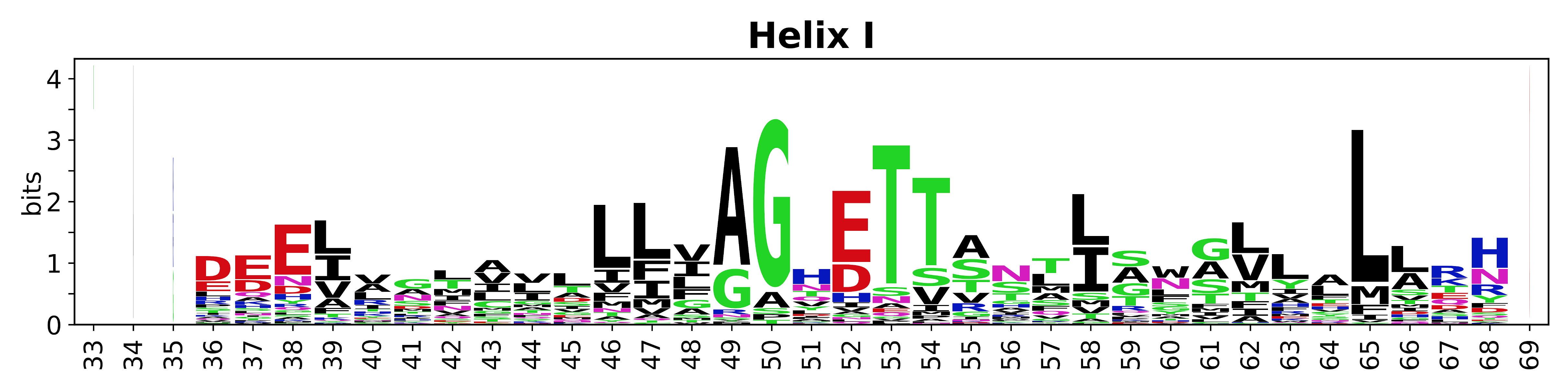
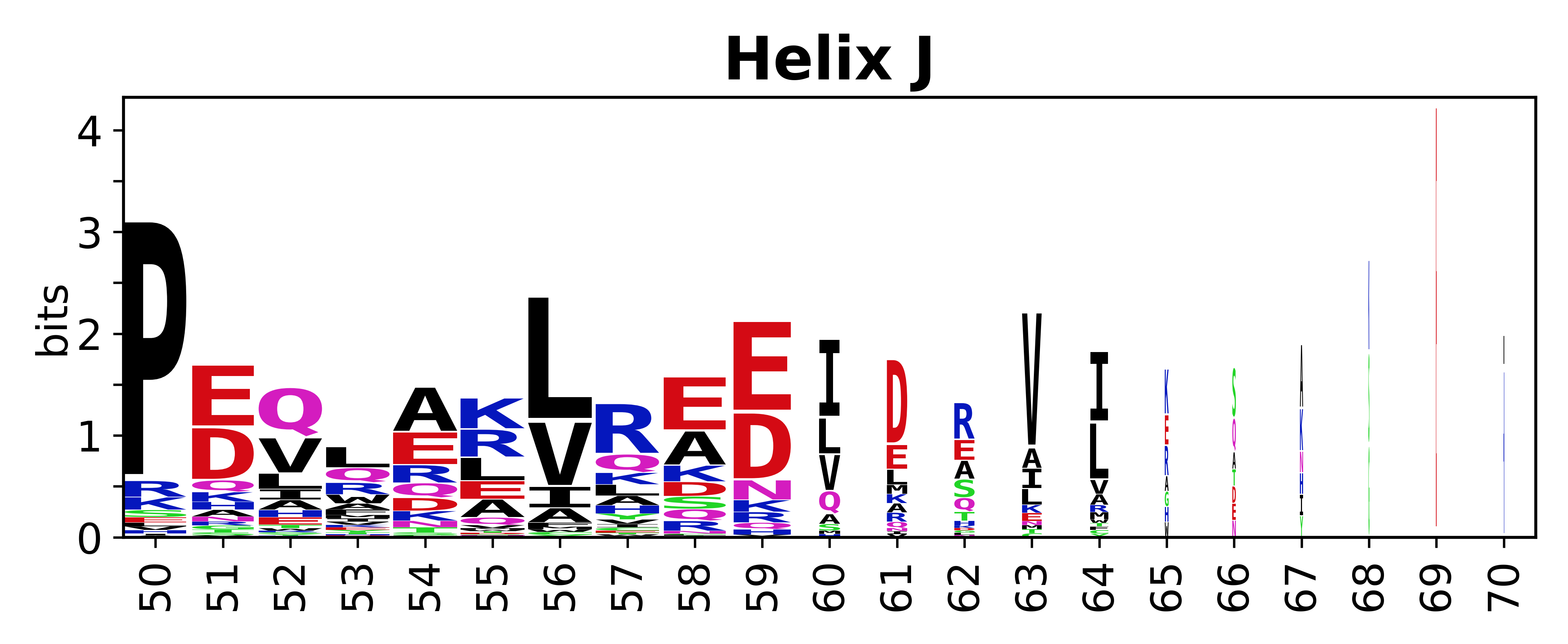
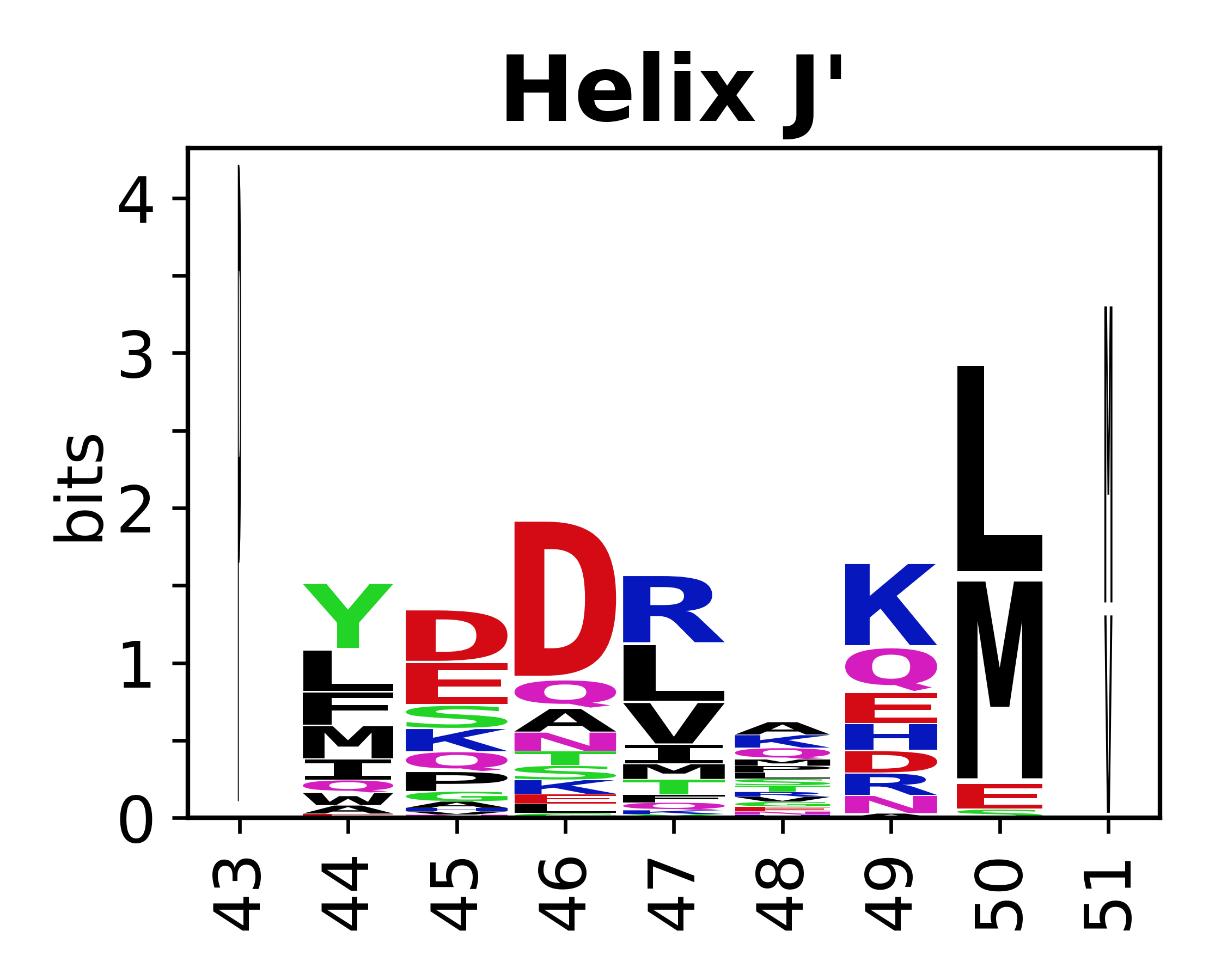
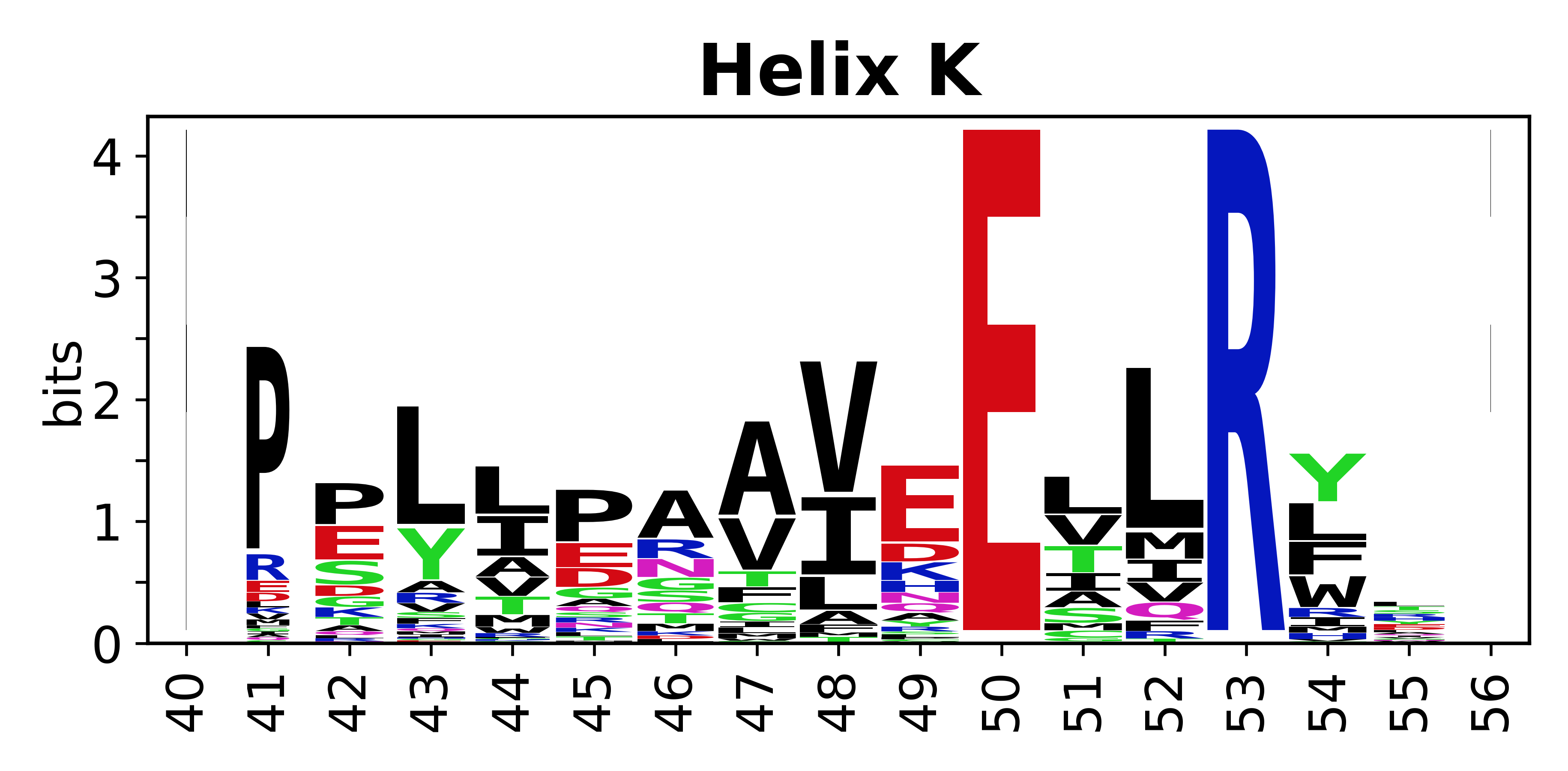
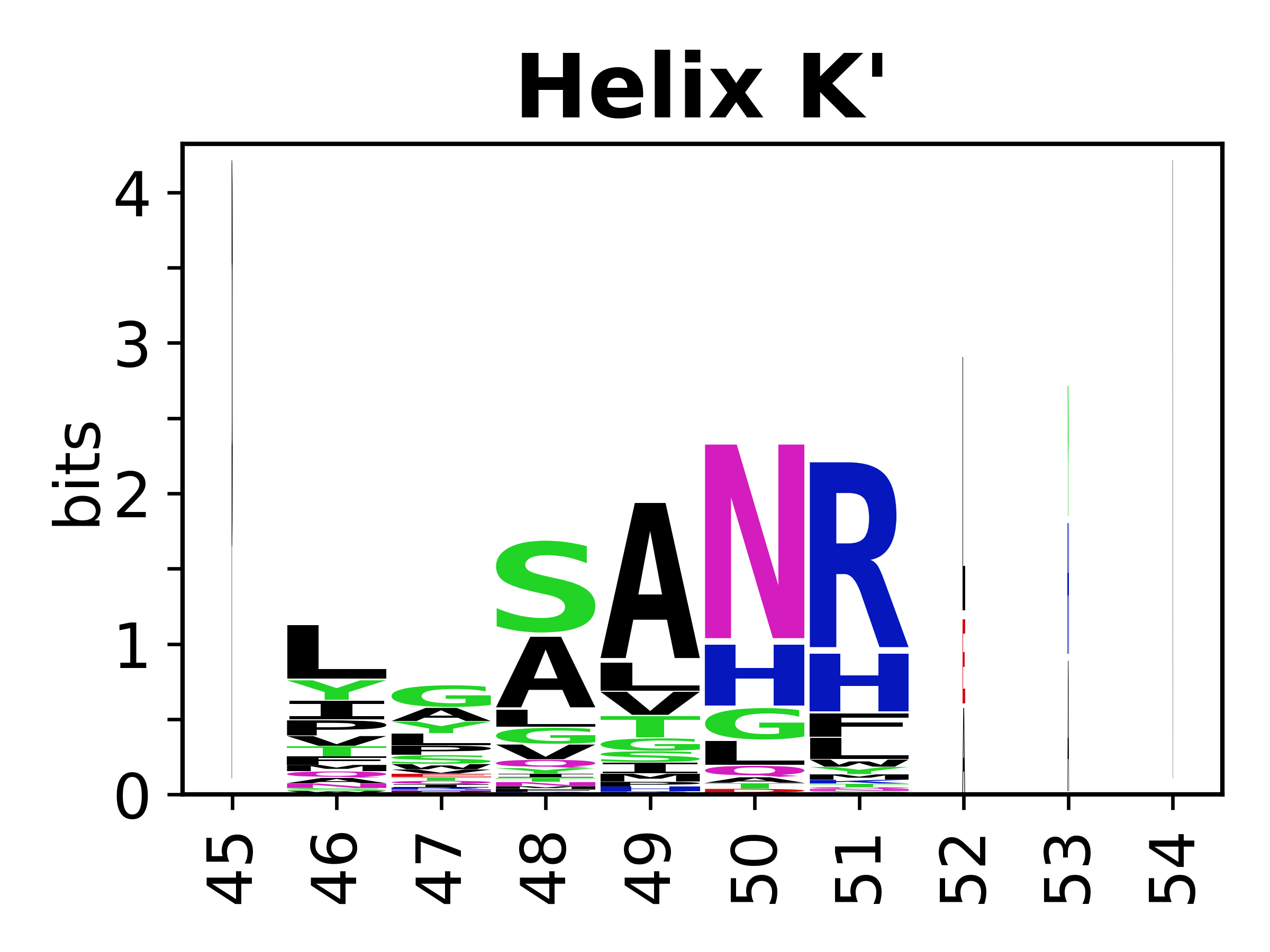
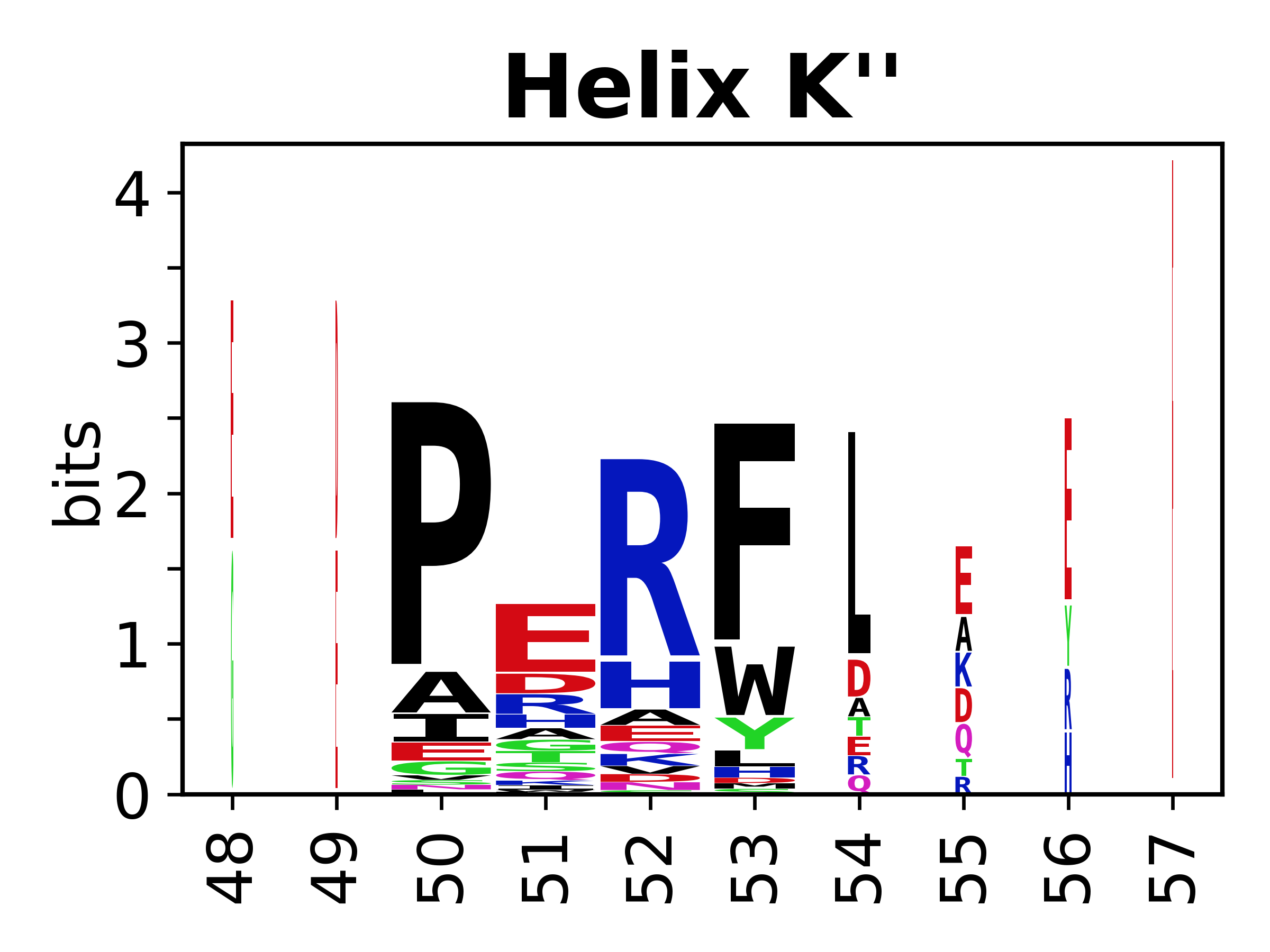
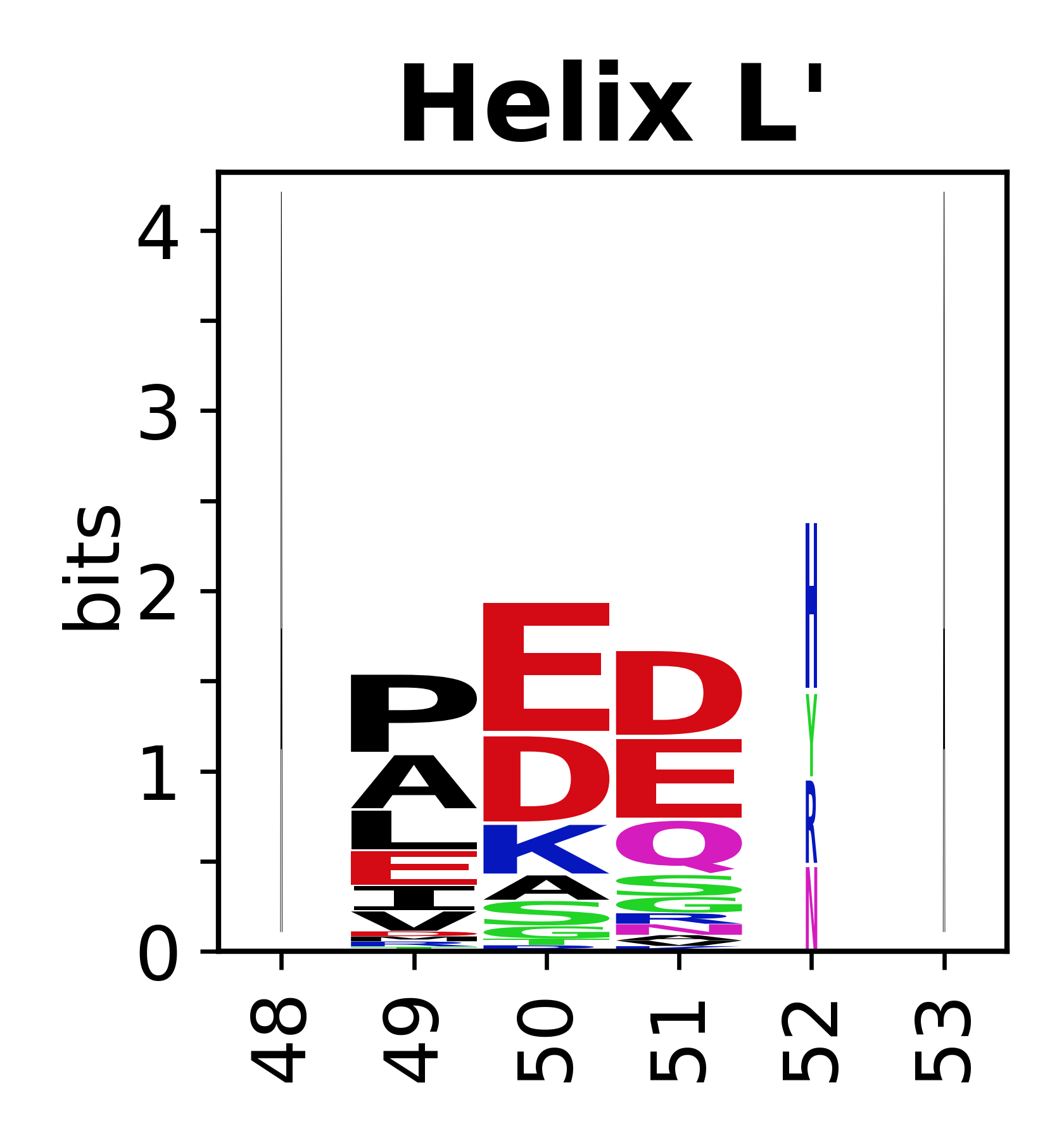
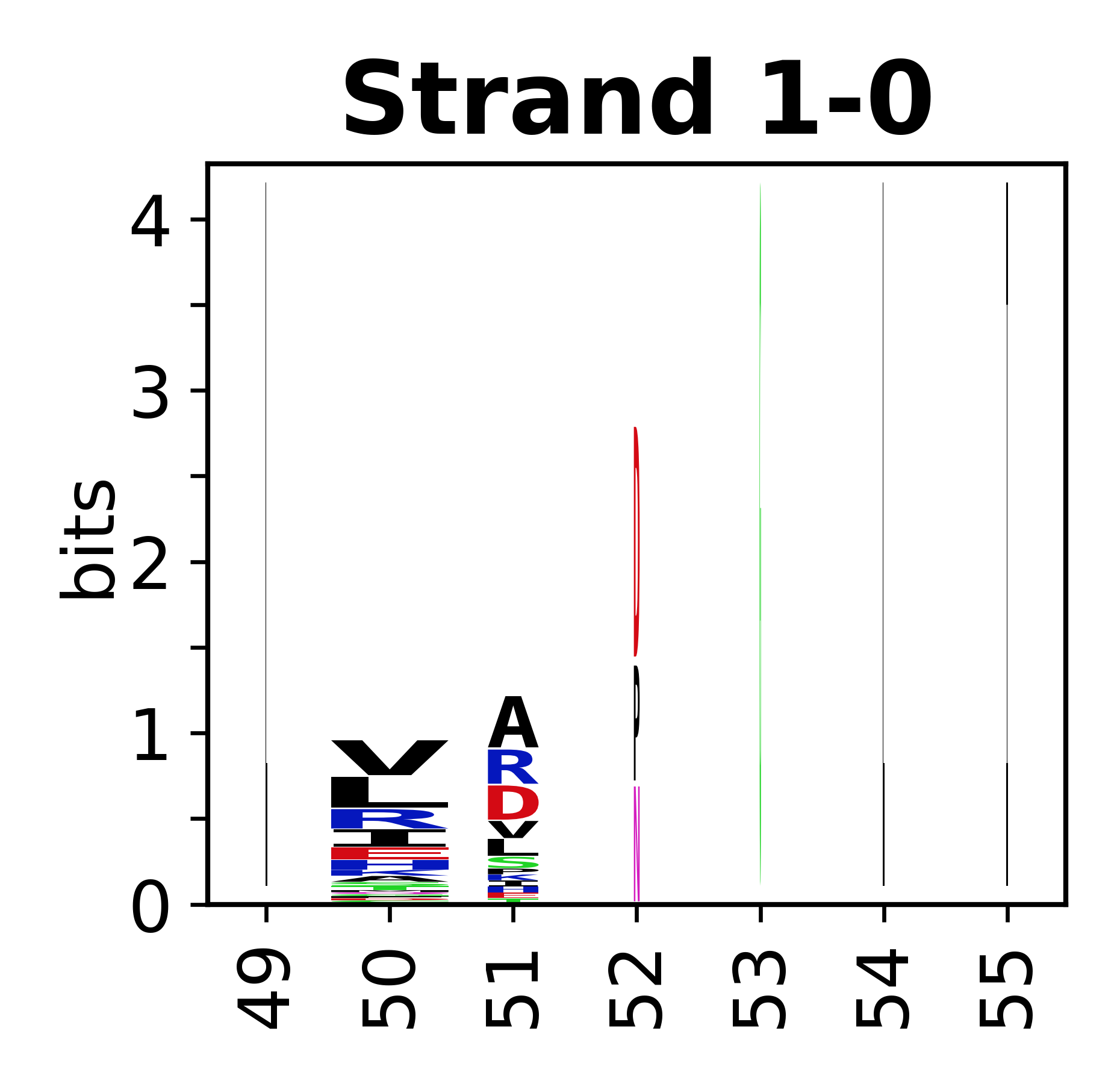
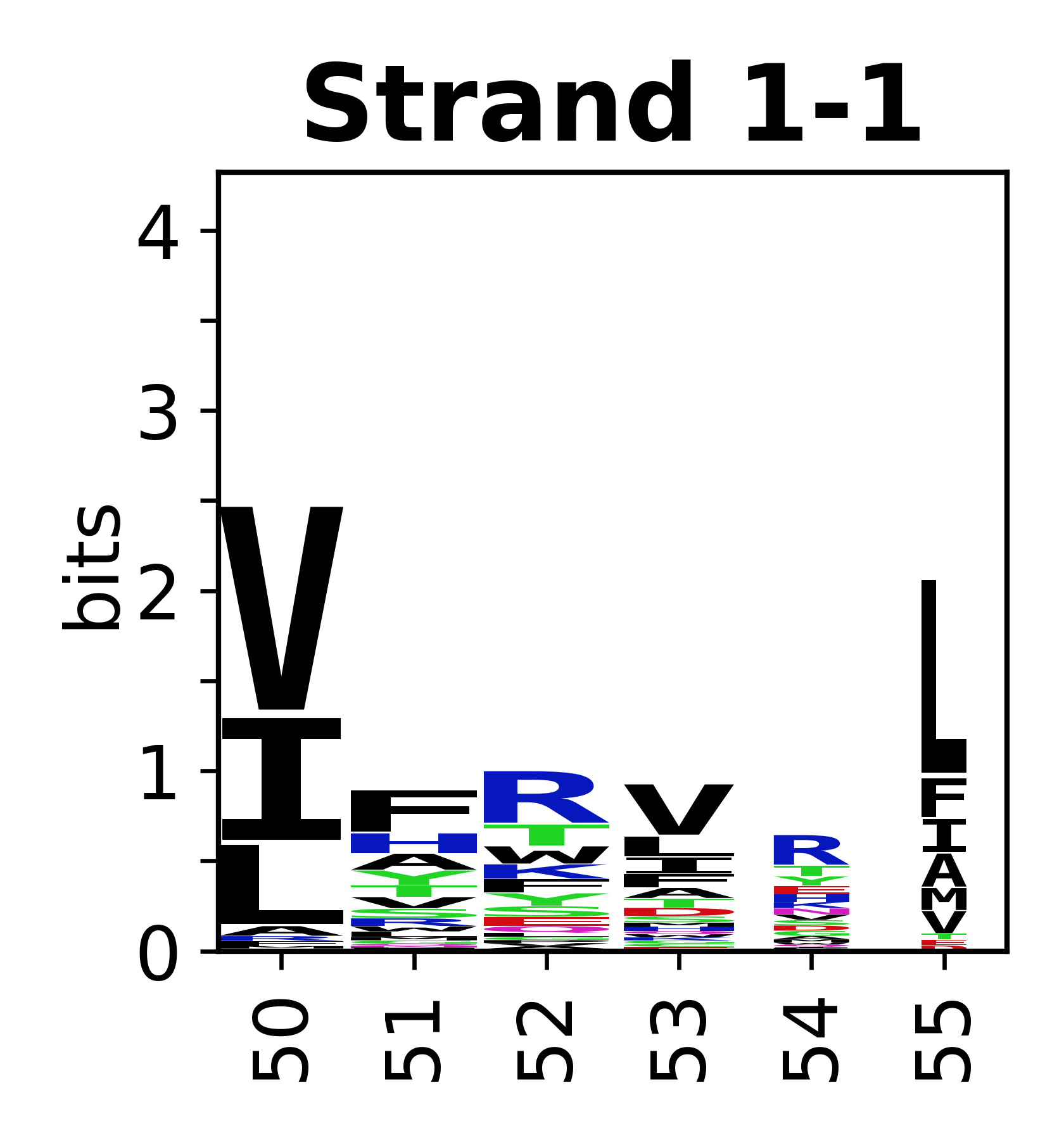
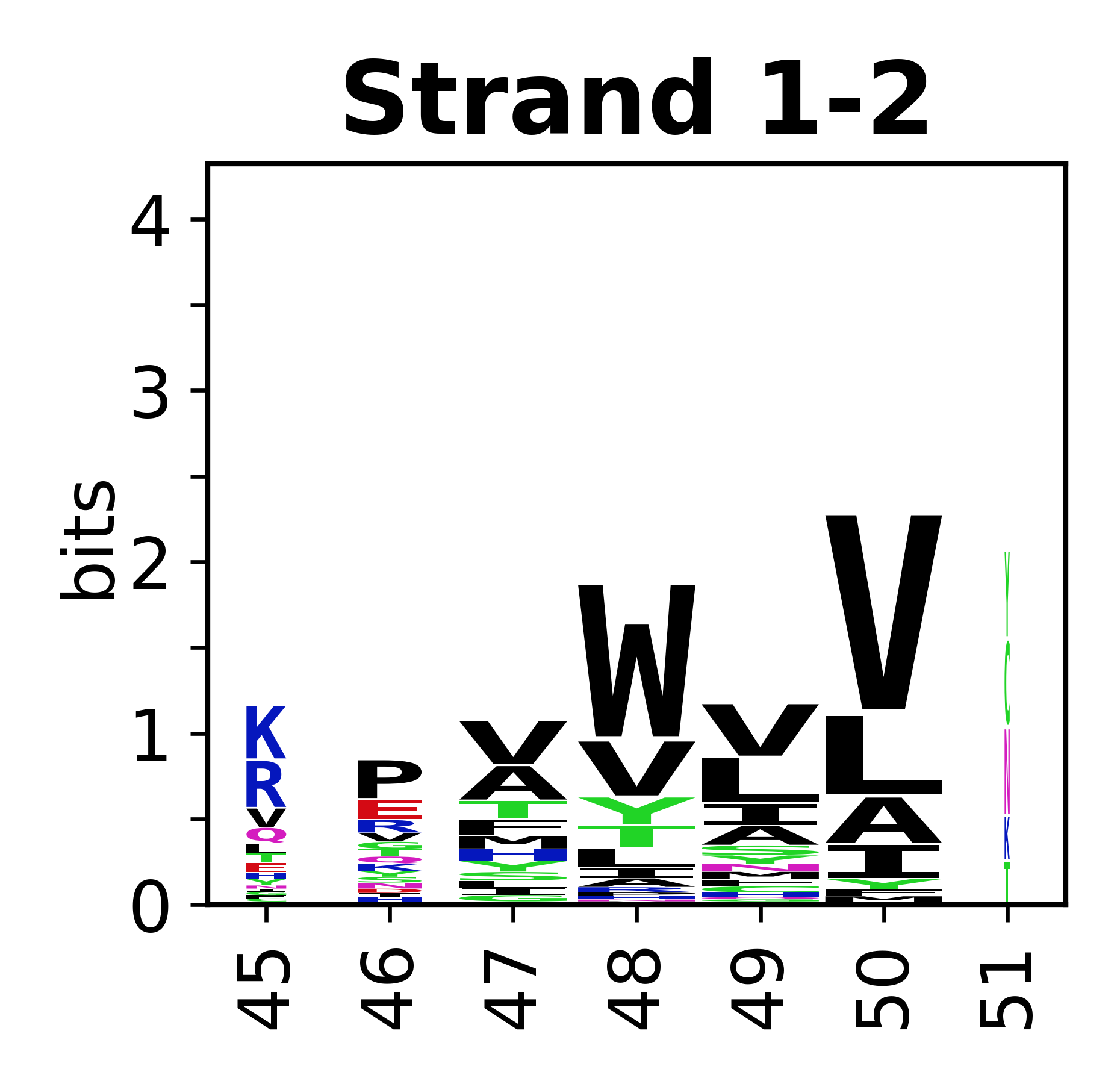
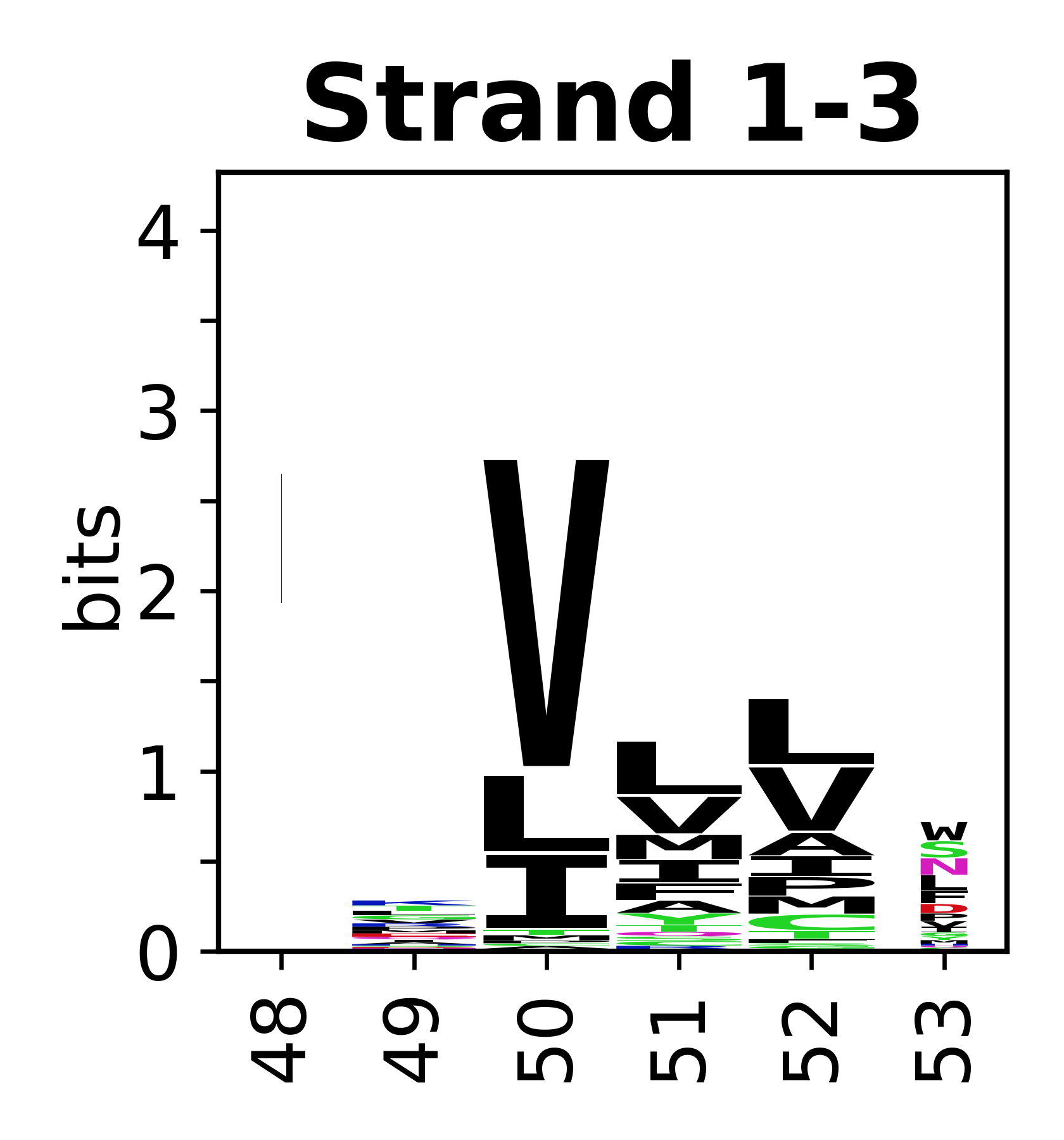
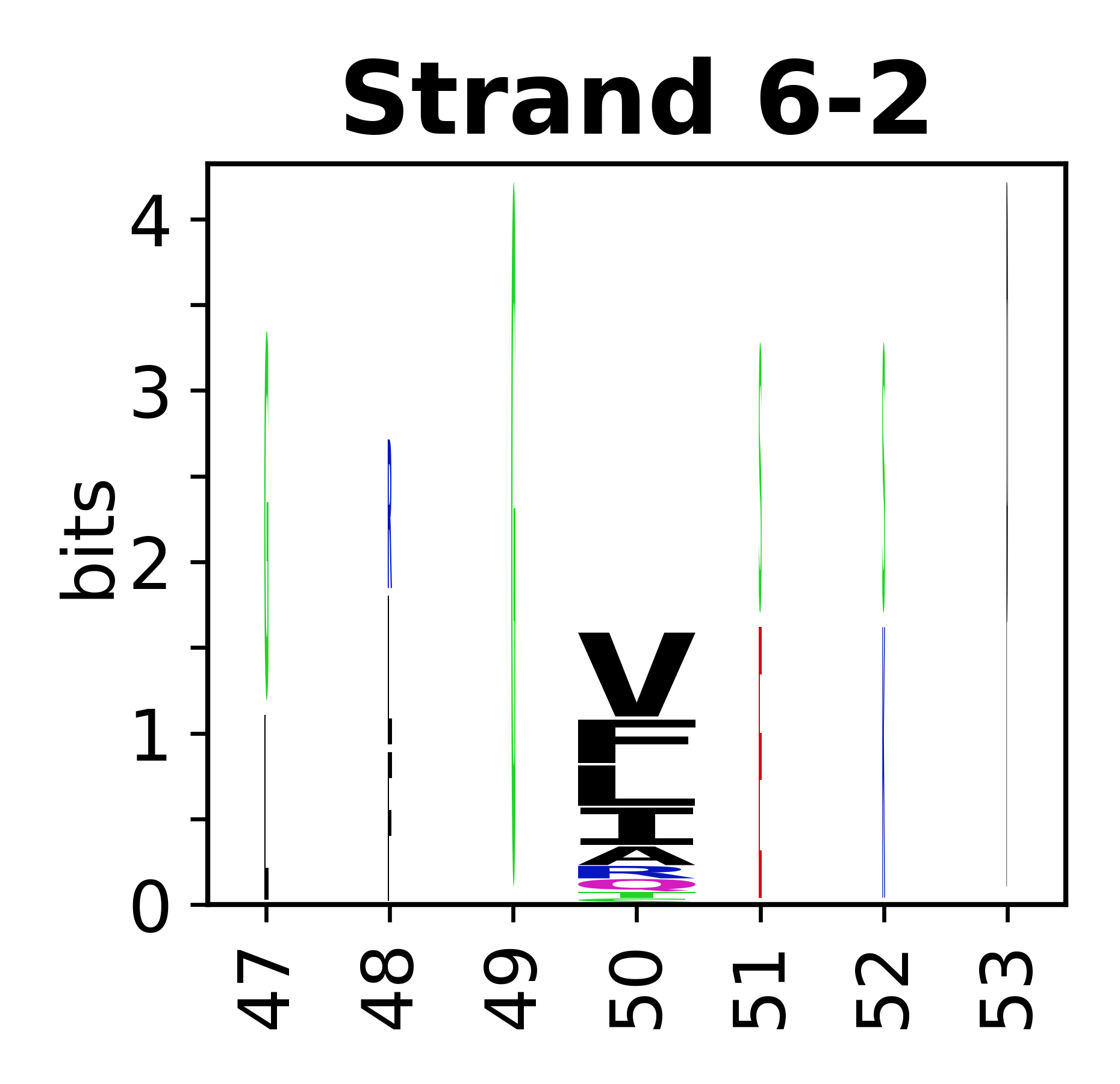
The amino acid sequences for each SSE can be aligned and used to produce a sequence logo. Where the sequence conservation is sufficient, we can establish a generic numbering scheme: the most conserved residue in helix X serves as its reference residue and is numbered as @X.50. The remaining residues in the helix are numbered accordingly. | The amino acid sequences for each SSE can be aligned and used to produce a sequence logo. Where the sequence conservation is sufficient, we can establish a generic numbering scheme: the most conserved residue in helix X serves as its reference residue and is numbered as @X.50. The remaining residues in the helix are numbered accordingly. | ||
<gallery mode="packed" heights= | <gallery mode="packed" heights=140px caption="Helices"> | ||
File:SecStrAnnotator-cyp-sse-logo-A-.png | File:SecStrAnnotator-cyp-sse-logo-A-.png | ||
File:SecStrAnnotator-cyp-sse-logo-A.png | File:SecStrAnnotator-cyp-sse-logo-A.png | ||
| Line 58: | Line 58: | ||
</gallery> | </gallery> | ||
<gallery mode="packed" heights= | <gallery mode="packed" heights=140px caption="Beta strands"> | ||
File:SecStrAnnotator-cyp-sse-logo-1-0.png | File:SecStrAnnotator-cyp-sse-logo-1-0.png | ||
File:SecStrAnnotator-cyp-sse-logo-1-1.png | File:SecStrAnnotator-cyp-sse-logo-1-1.png | ||
Revision as of 15:49, 20 March 2020
SecStrAnnotator Suite provides scripts (Python, R, and bash) for batch annotation of the whole family and analysis of the annotation results. The main scripts are
- SecStrAPI_master.sh – this bash script used to prepare data for SecStrAPI also formats the annotations into tab-separated (TSV) files readable by R
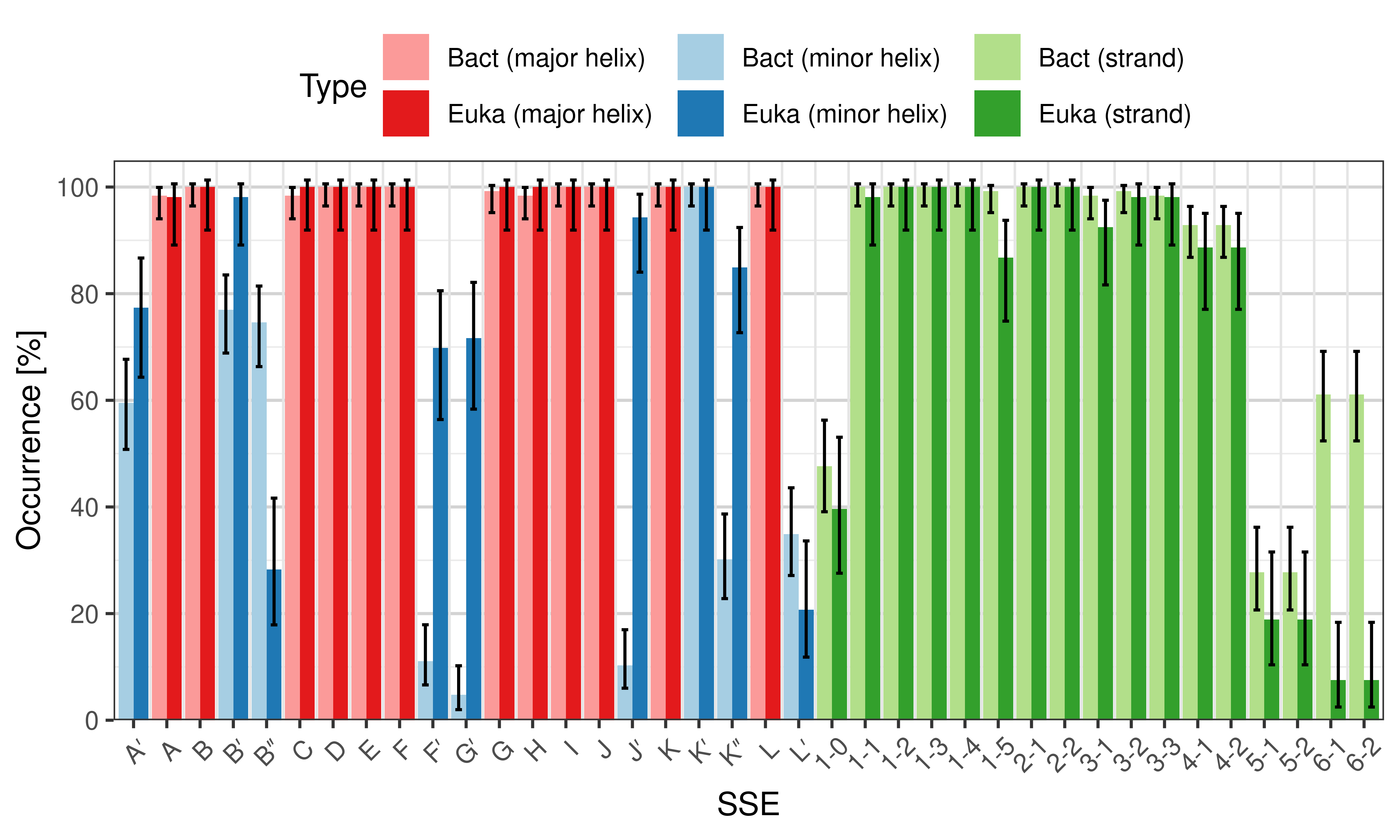
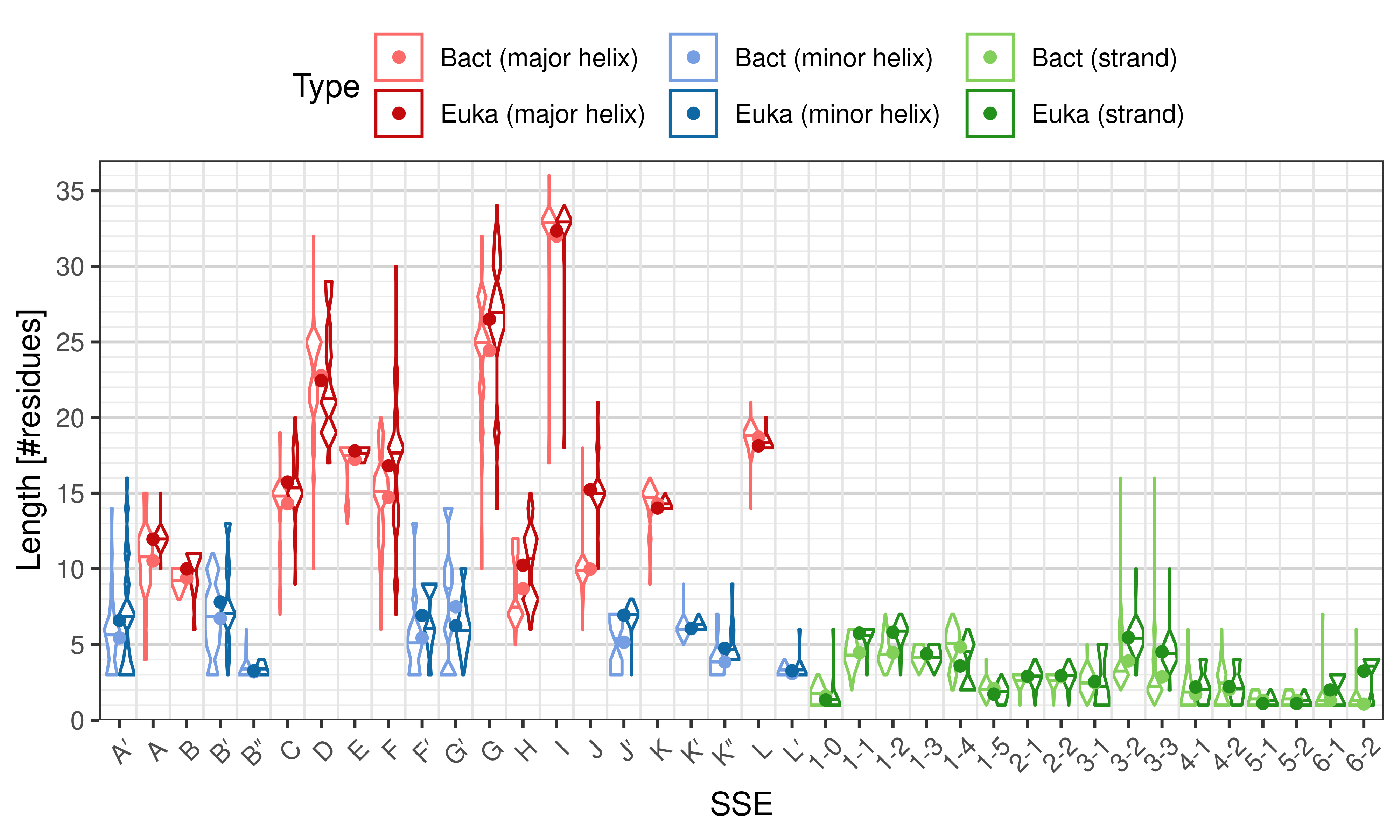
- secondary_structure_anatomy.R – contains commands for reading the annotation results, generating plots, and performing some statistical test to compare eukaryotic and bacterial structures (or any two sets of structures)
Example case study: Cytochromes P450
Data
For the Cytochrome P450 family, structures of 1775 protein domains are available, located in 953 PDB entries. The analysis was performed on a non-redundant subset containing 175 protein domains.
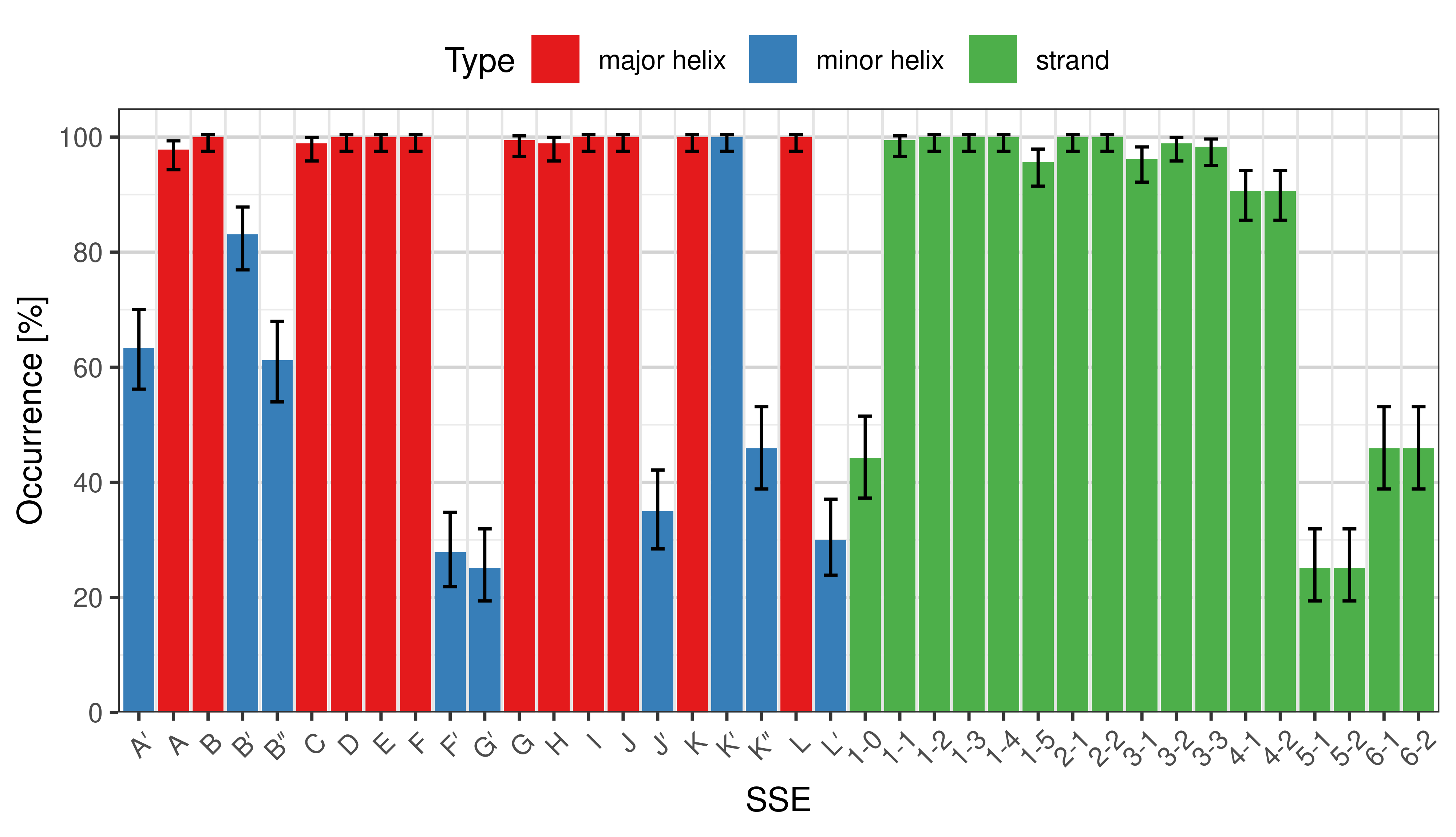
Occurrence of SSEs
The occurrence describes in what percentage of the structures a particular SSE is present.
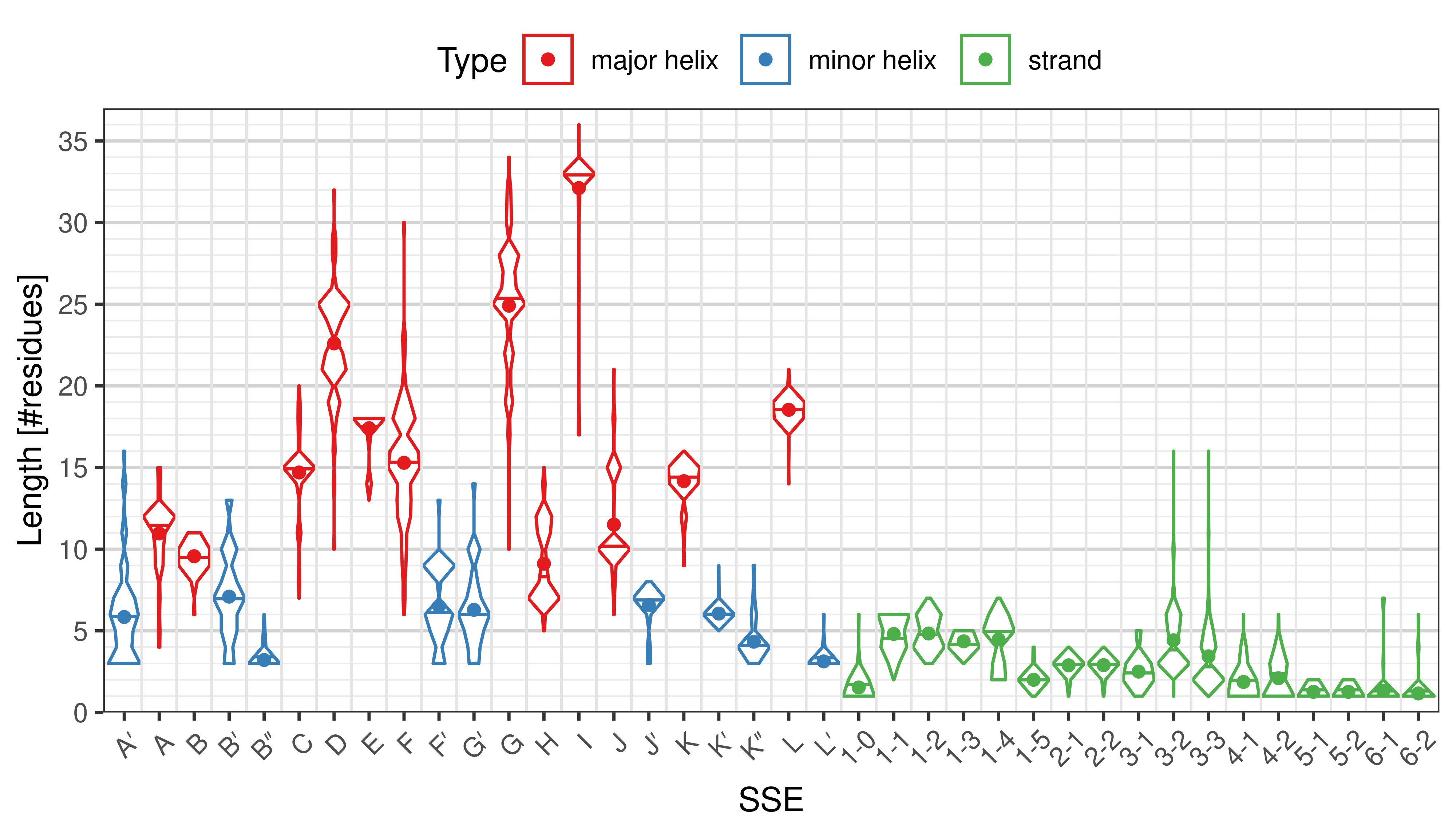
Length of SSEs
The length of an SSE is measured as the number of residues. The following violin plots show the distribution of length for each SSE.
Sequence of SSEs
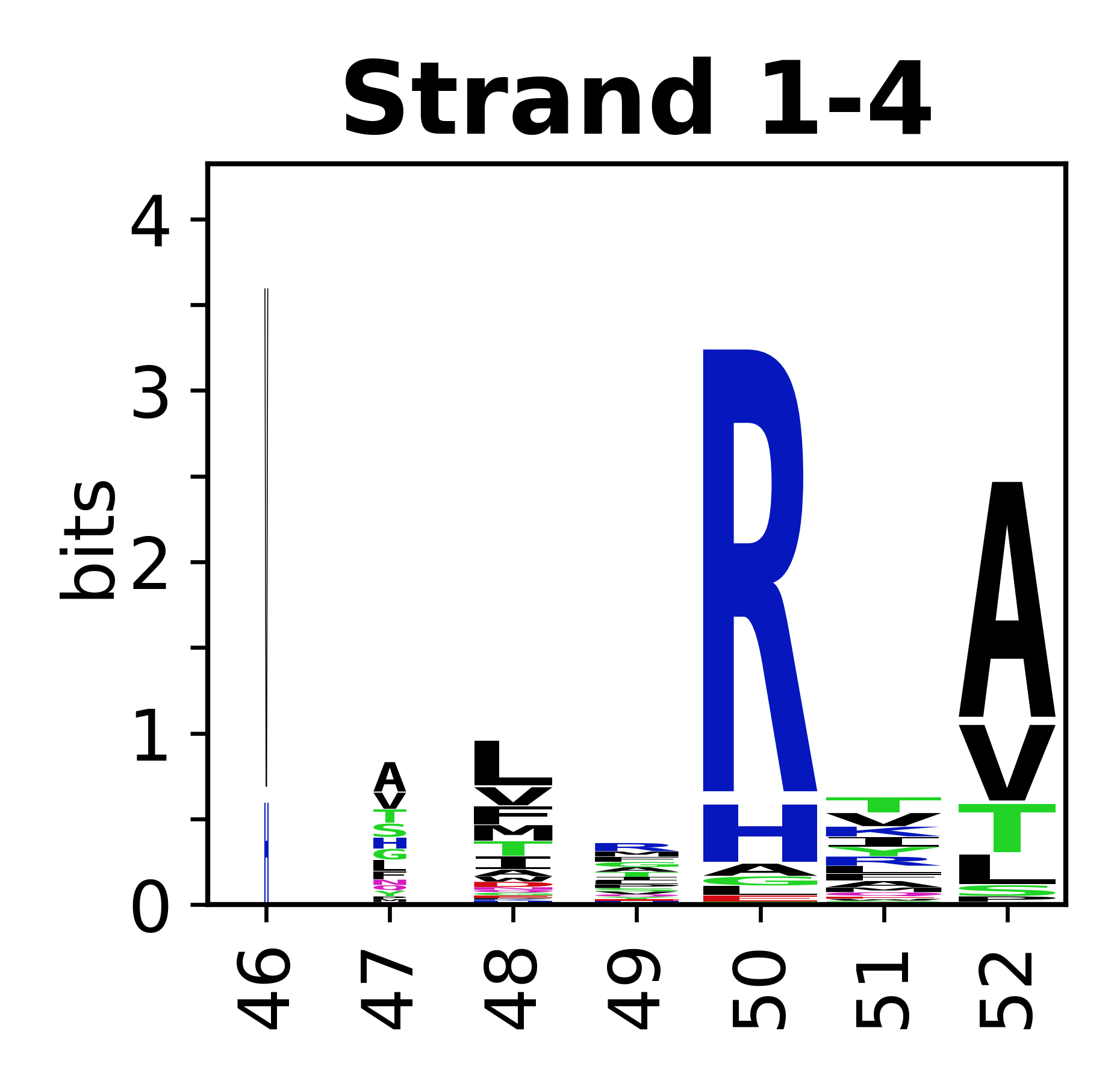
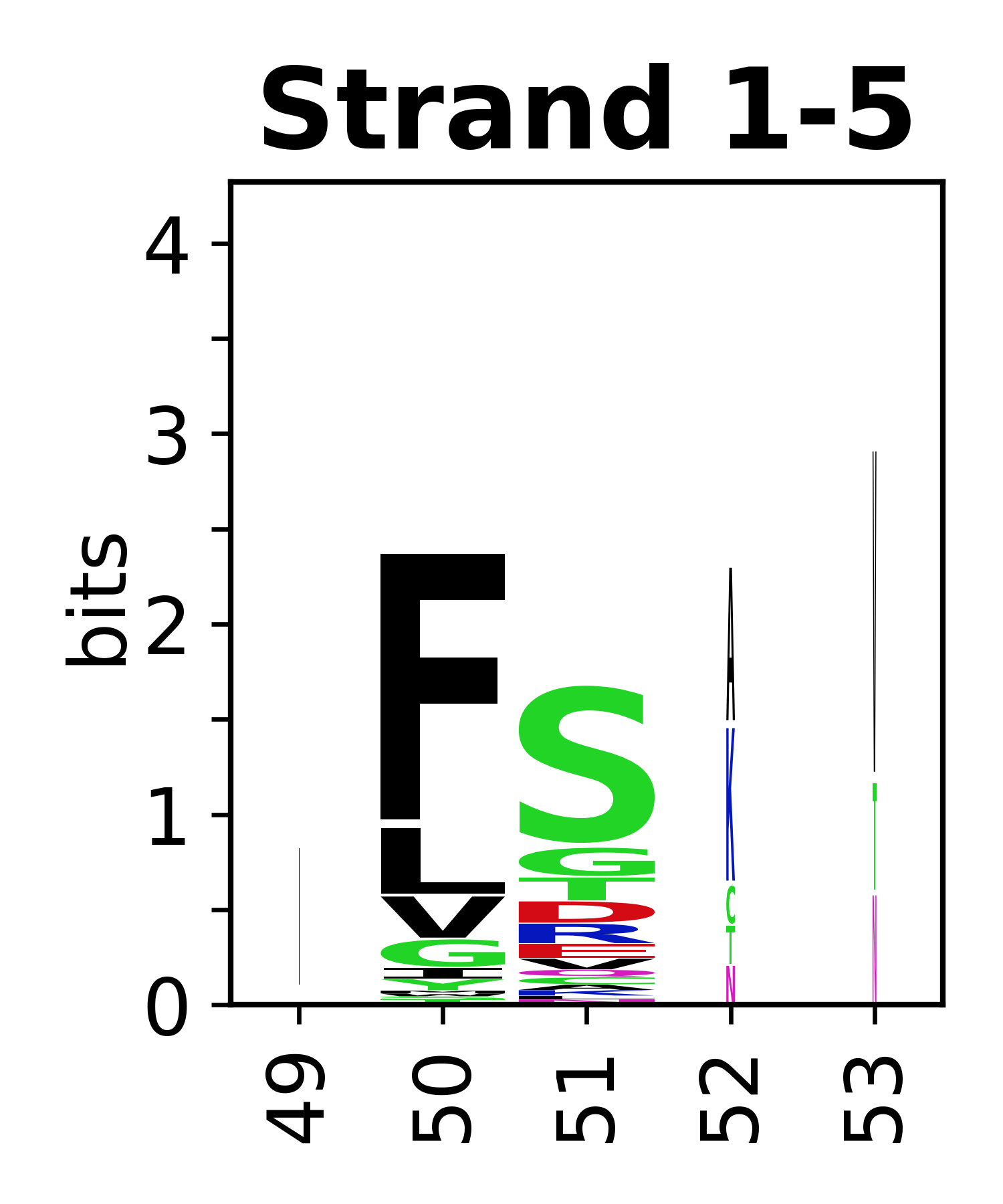
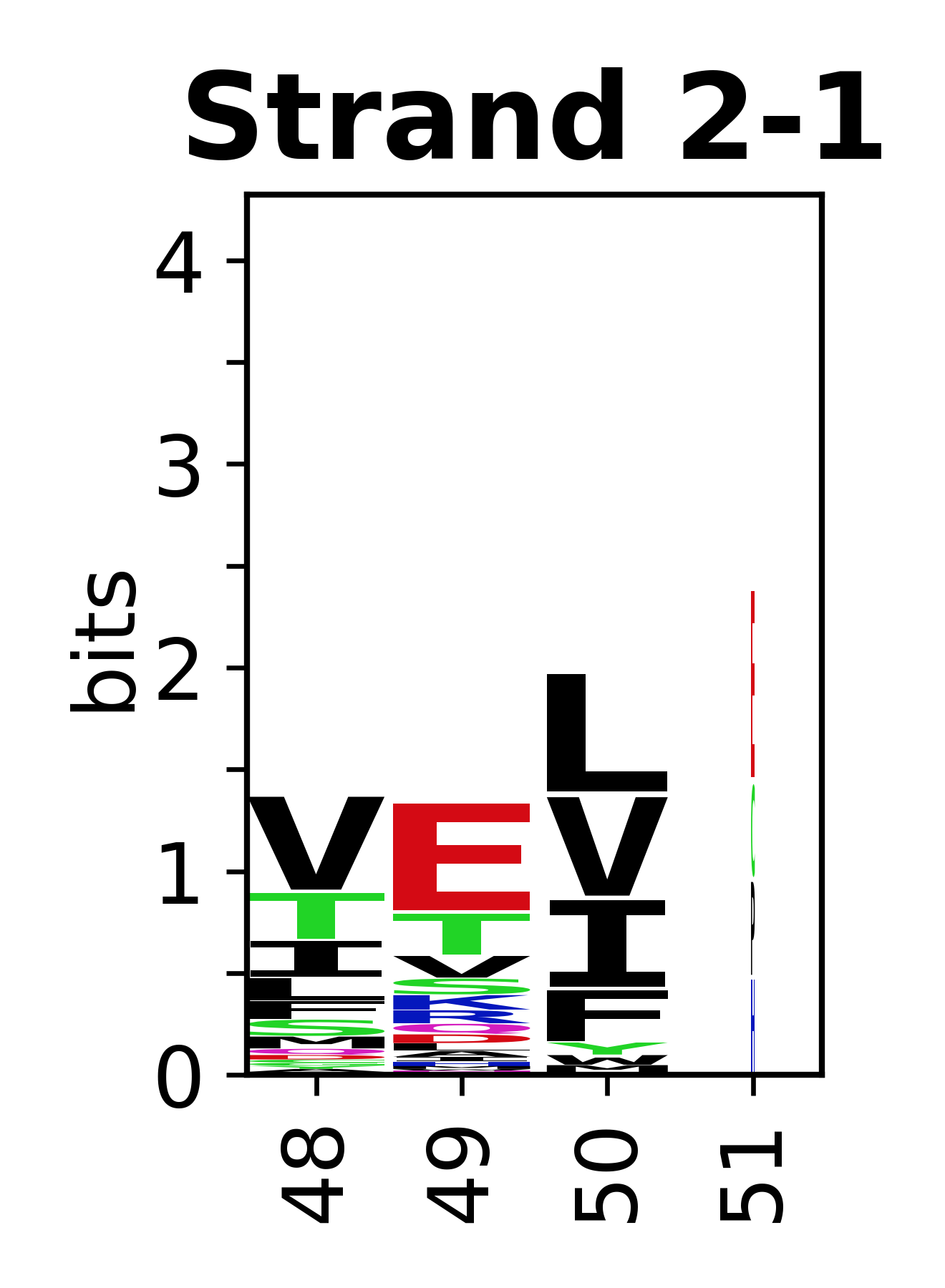
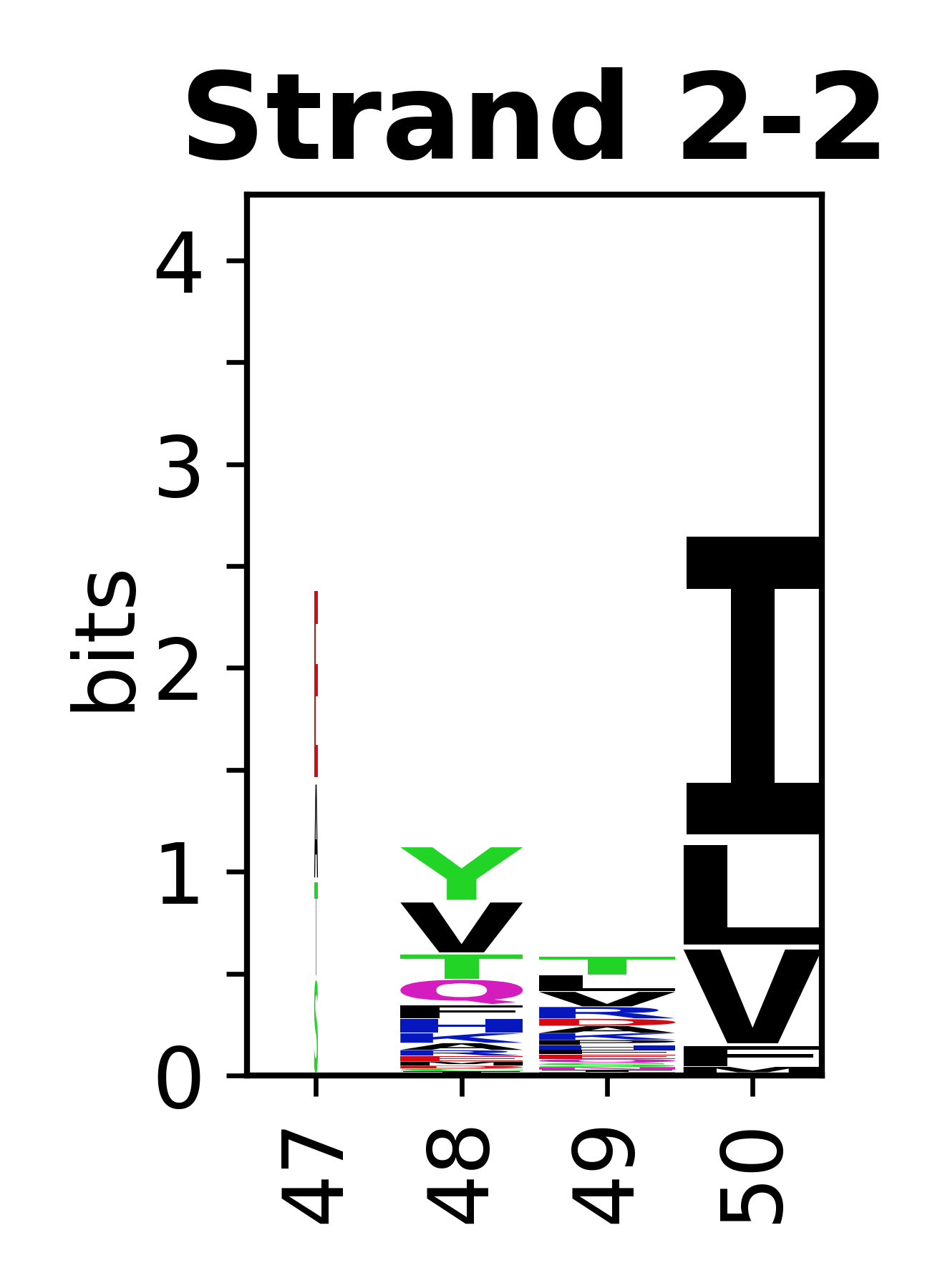
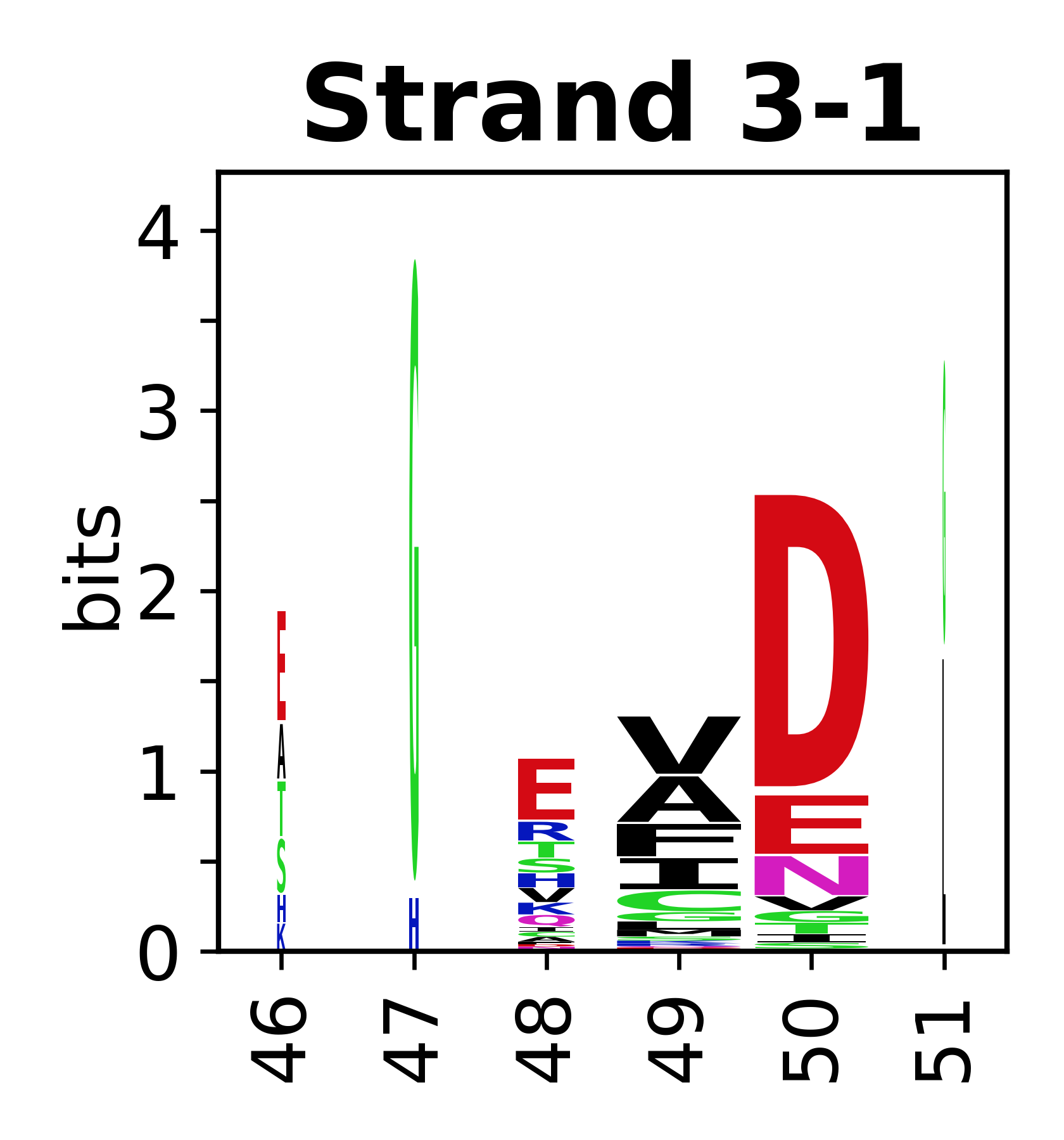
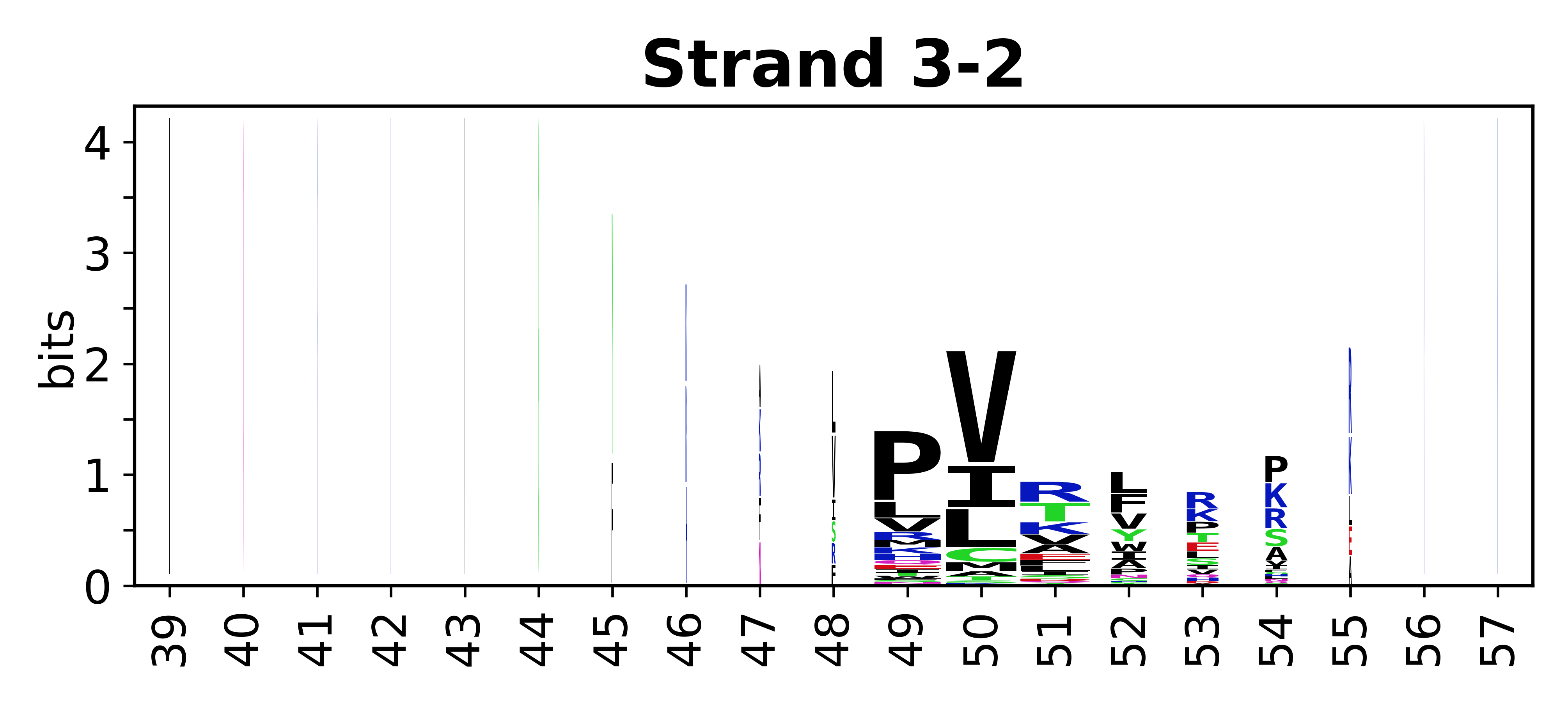
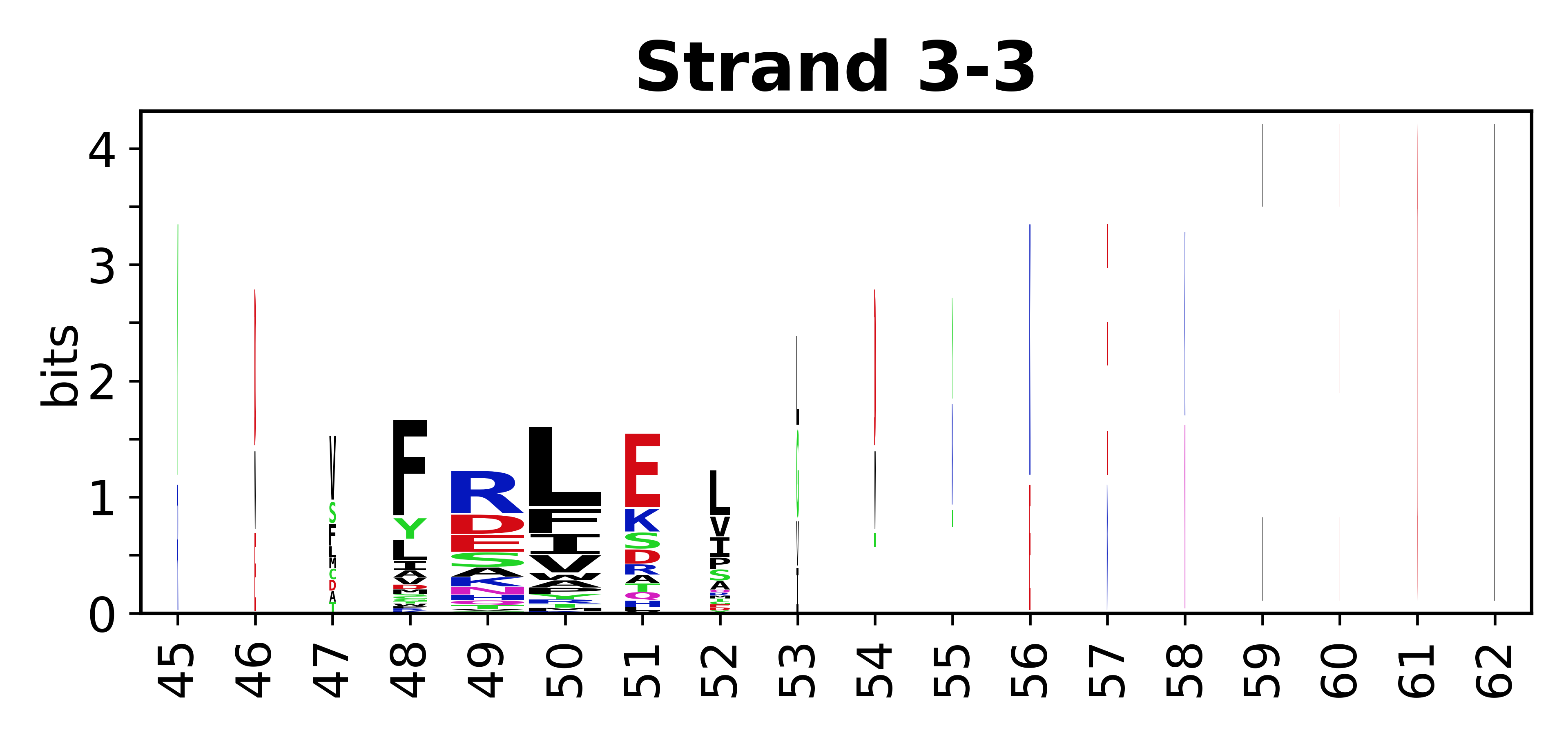
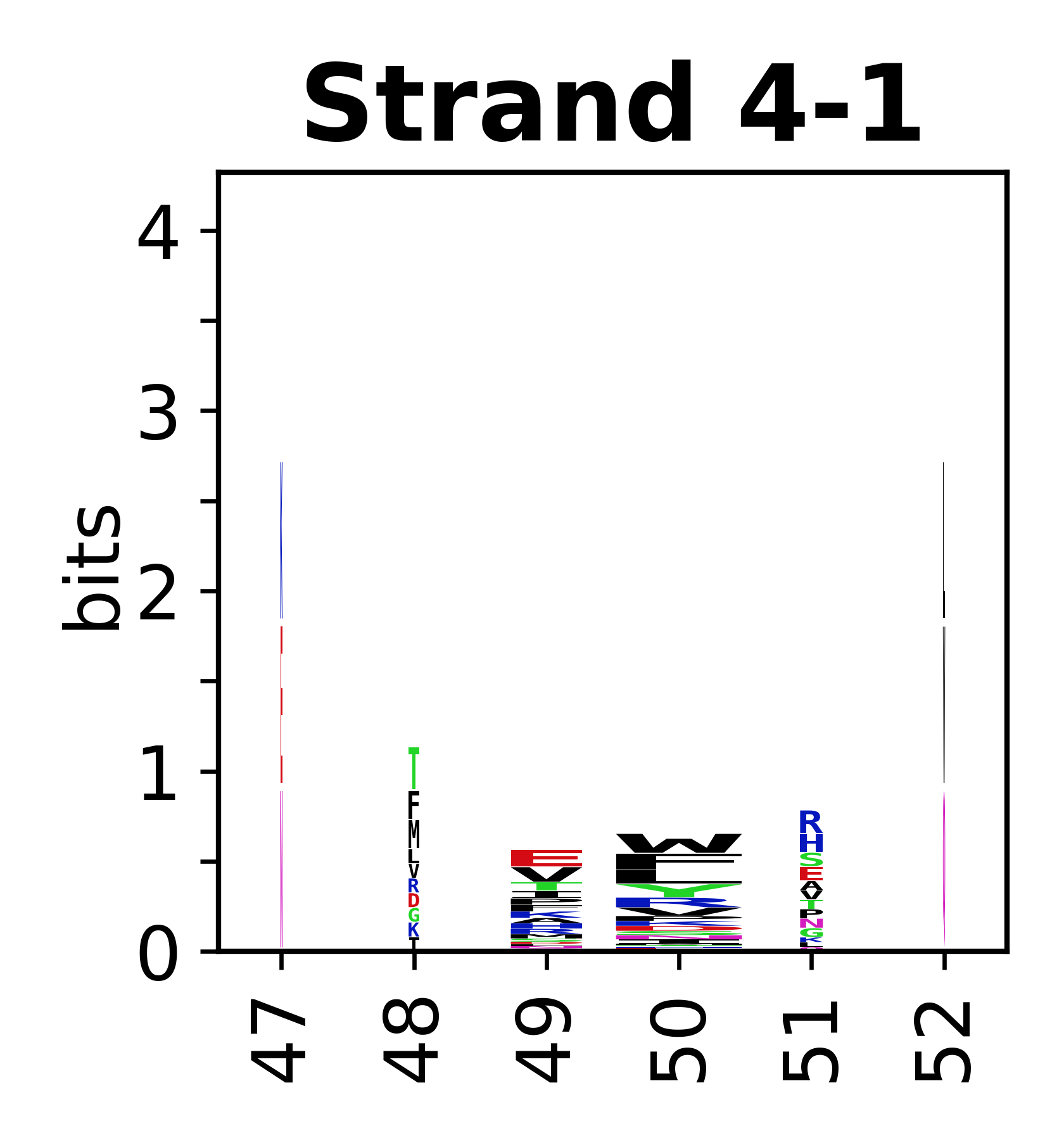
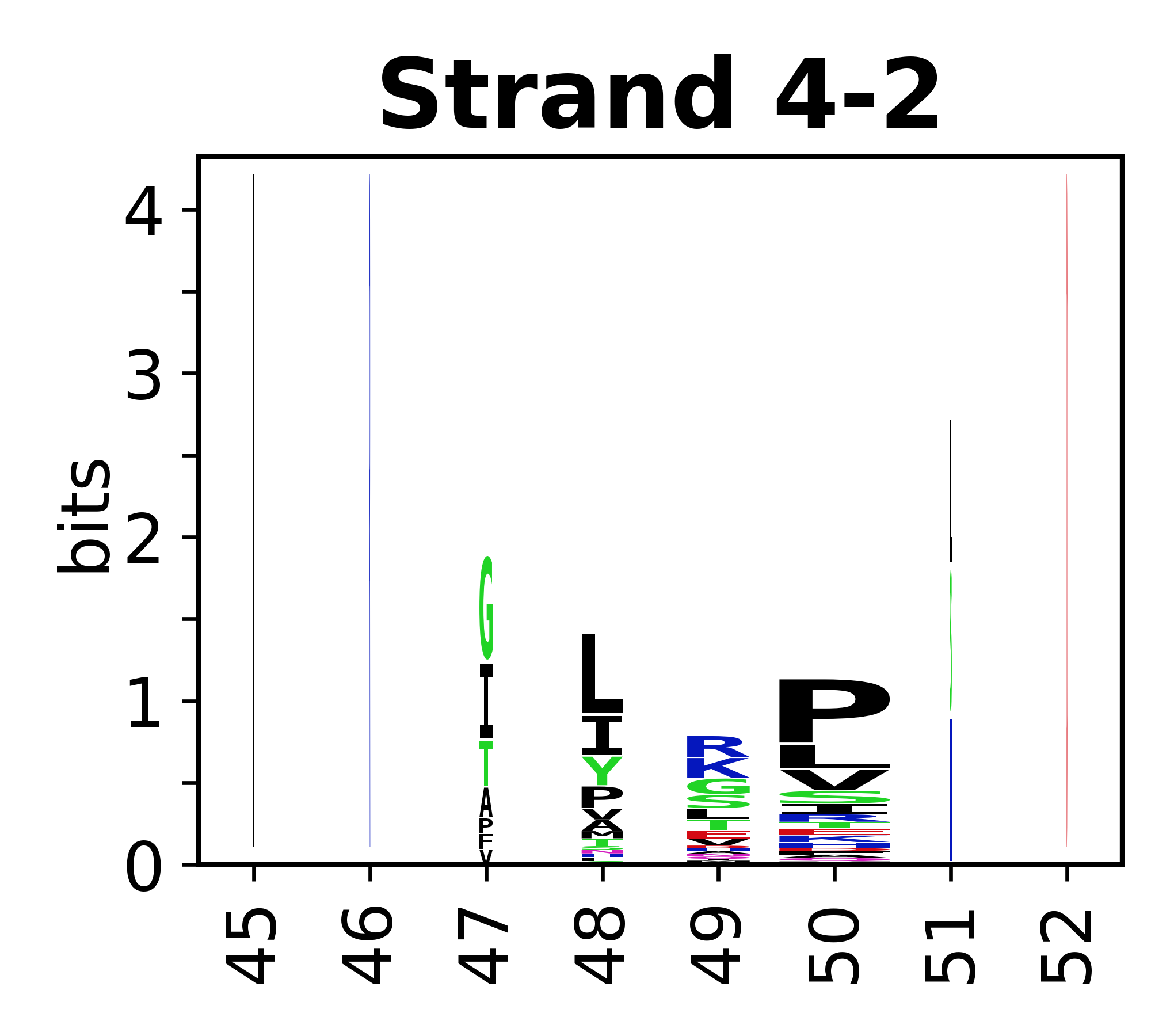
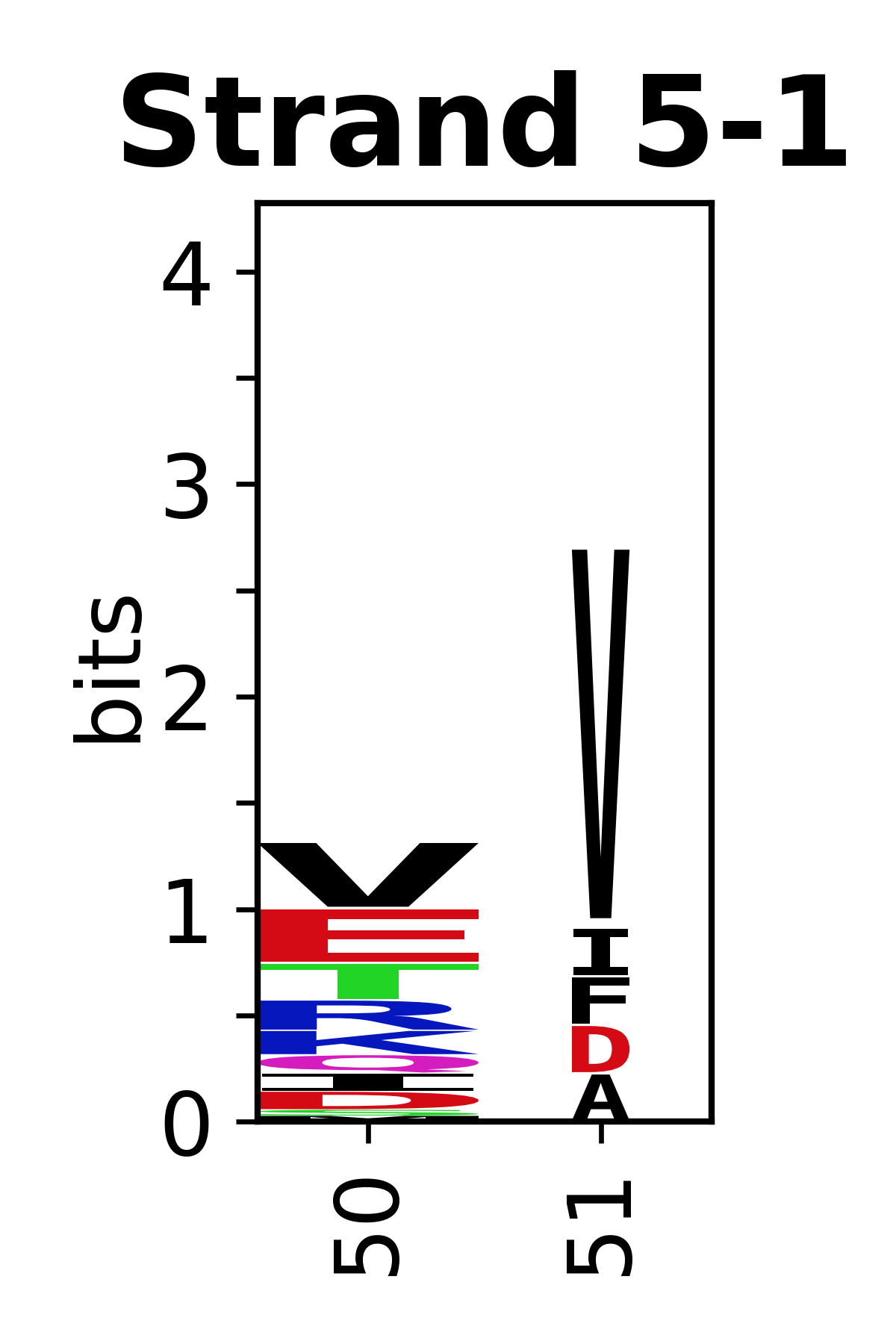
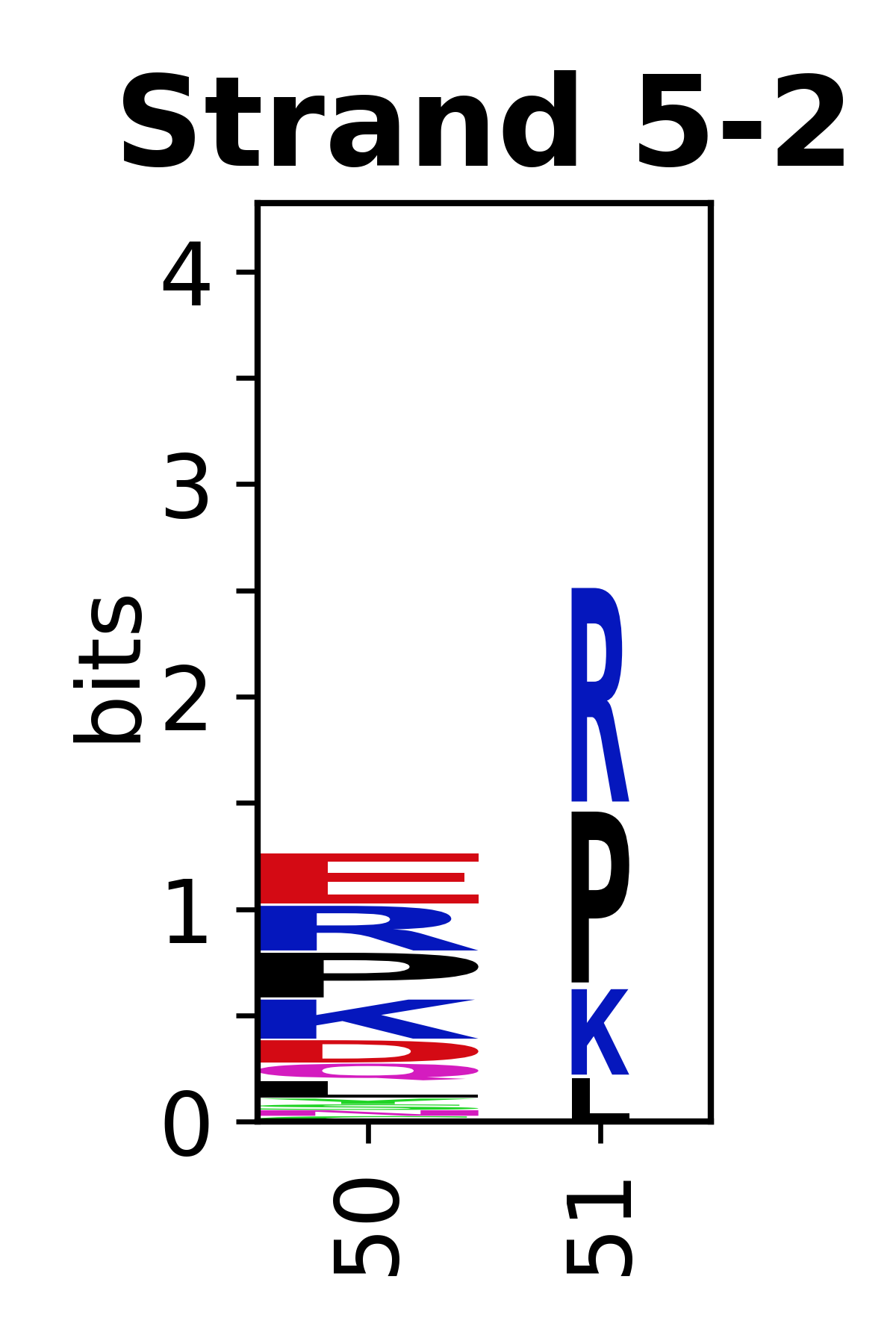
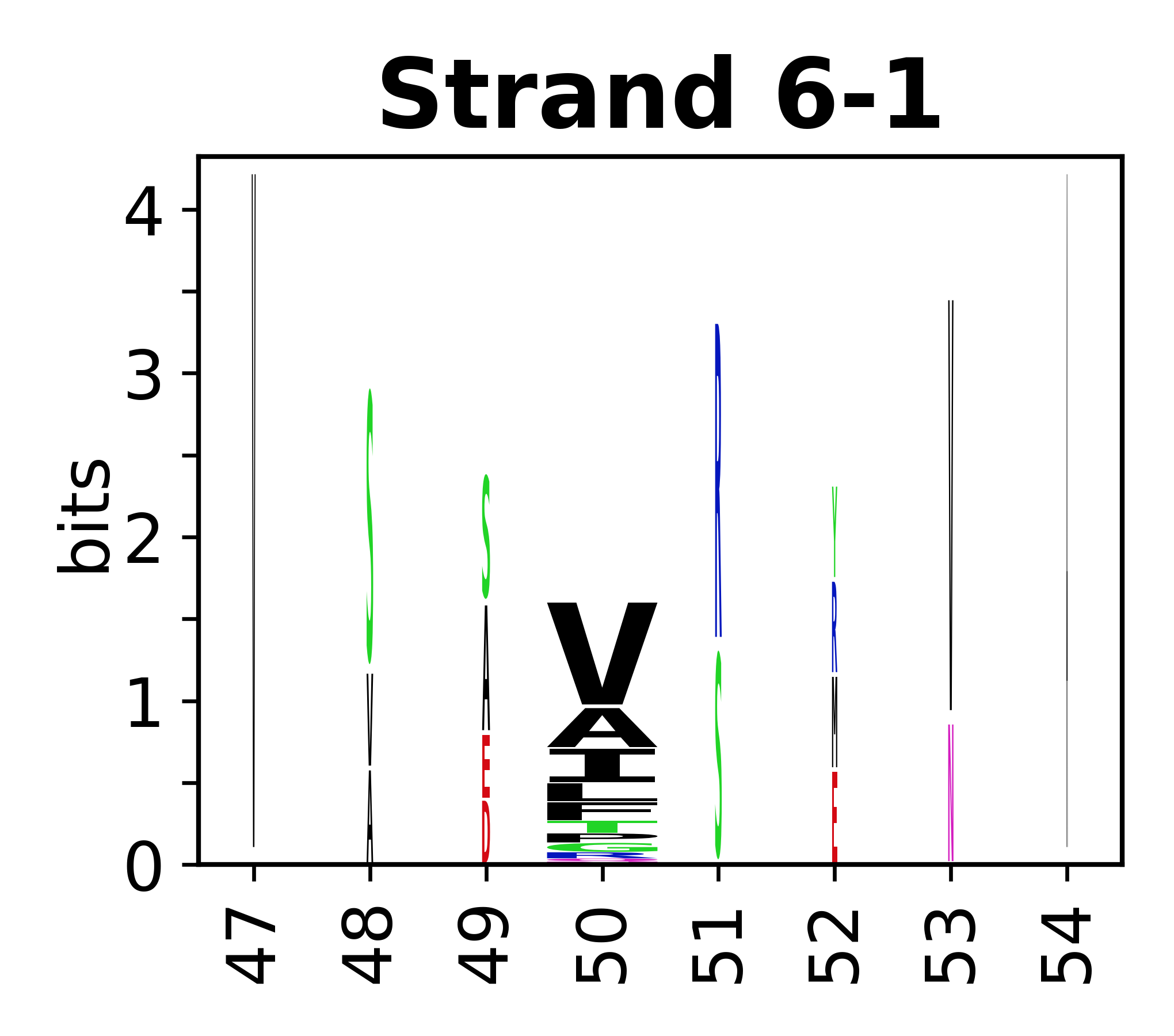
The amino acid sequences for each SSE can be aligned and used to produce a sequence logo. Where the sequence conservation is sufficient, we can establish a generic numbering scheme: the most conserved residue in helix X serves as its reference residue and is numbered as @X.50. The remaining residues in the helix are numbered accordingly.
- Helices
- Beta strands