NEEMP:Reports: Difference between revisions
Appearance
No edit summary |
No edit summary |
||
| Line 49: | Line 49: | ||
|- | |- | ||
|colspan=2|'''''Figure 8:''''' | |colspan=2|'''''Figure 8:''''' | ||
|} | |||
| } | |||
Revision as of 15:53, 29 June 2016
Along with NEEMP we provide an handy python script to generate charge correlation graphs and quality assay reports, named nut-report.py.
SCRIPT BASIC USAGE:
Step 1:
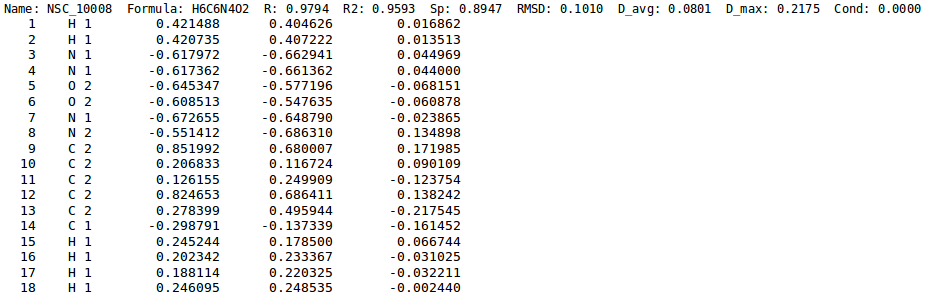
- Generation of chg-stats-out-file in which for each atom the difference between the reference QM charge and the EEM charge is calculated (figure 5). Such a file can be obtained running NEEMP in quality validation mode with the option
--chg-stats-out-file(see example or quality validation for details).
Step 2:
- Call nut-report.py with the above-mentioned file
./nut-report.py chg-stats-out-file
Requirements:
- Python (https://www.python.org/)
NB: the script has been tested for both Python 2.7 and Python 3.4, as well as for R 3.2.
.
Once the script has been executed, a bunch of output files are generated in the chg-stats-out-file directory. In particular:
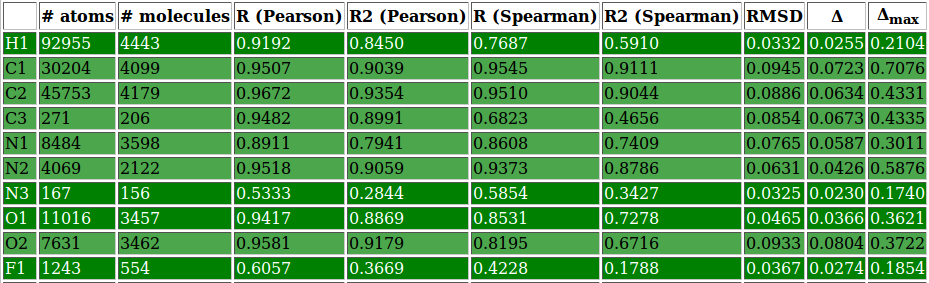
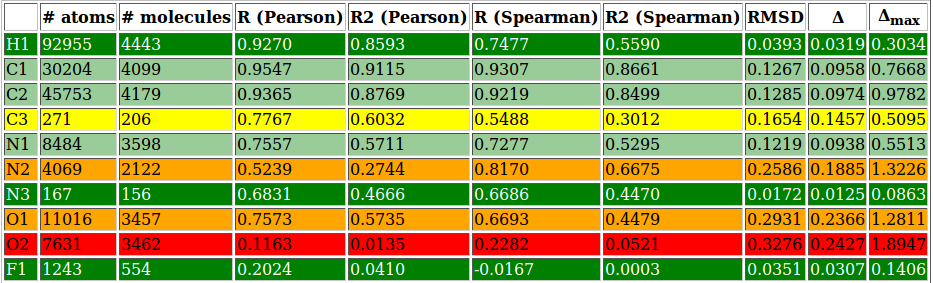
- csv files containing per-atom charge information and values for several performance evaluating metrics for each atomic type and molecule
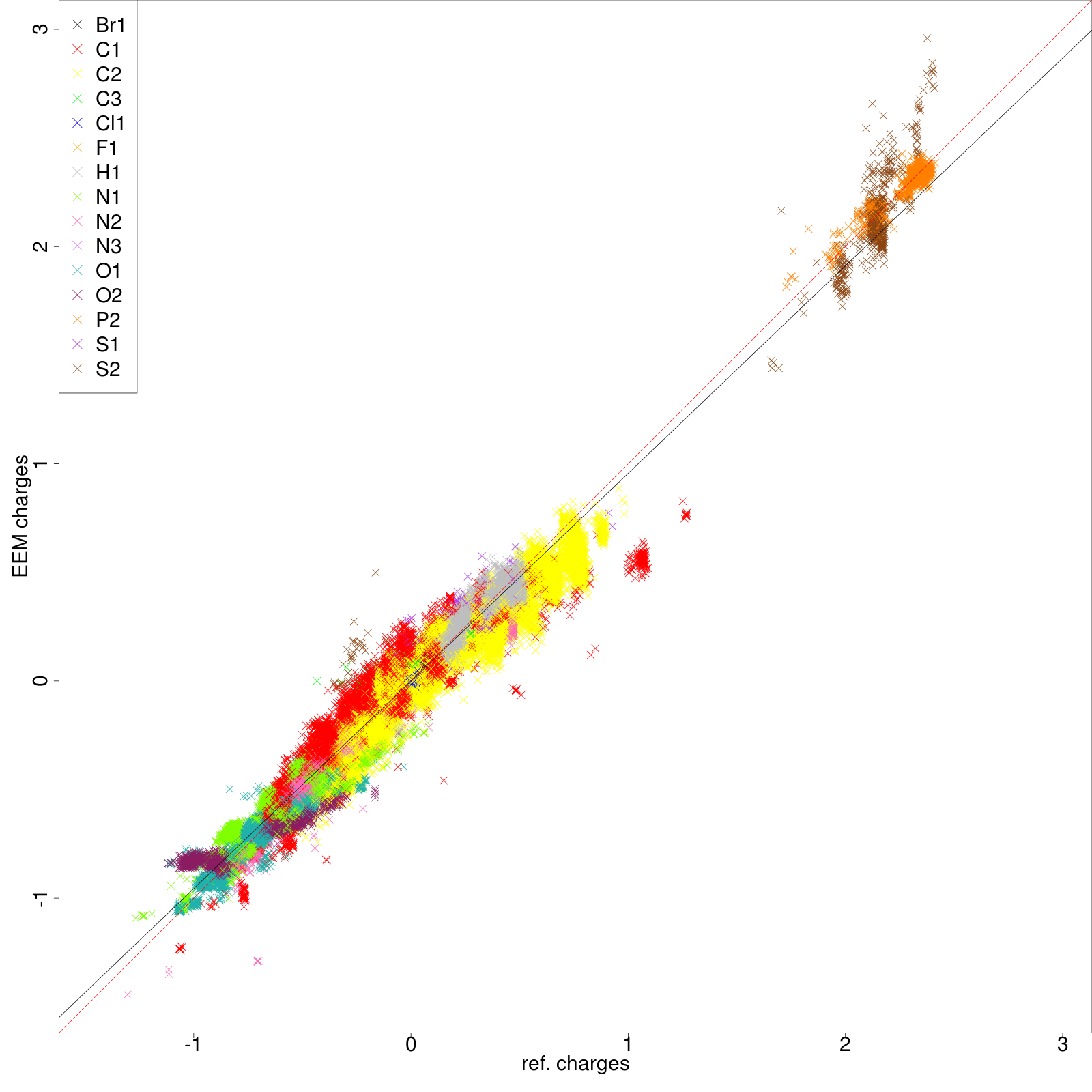
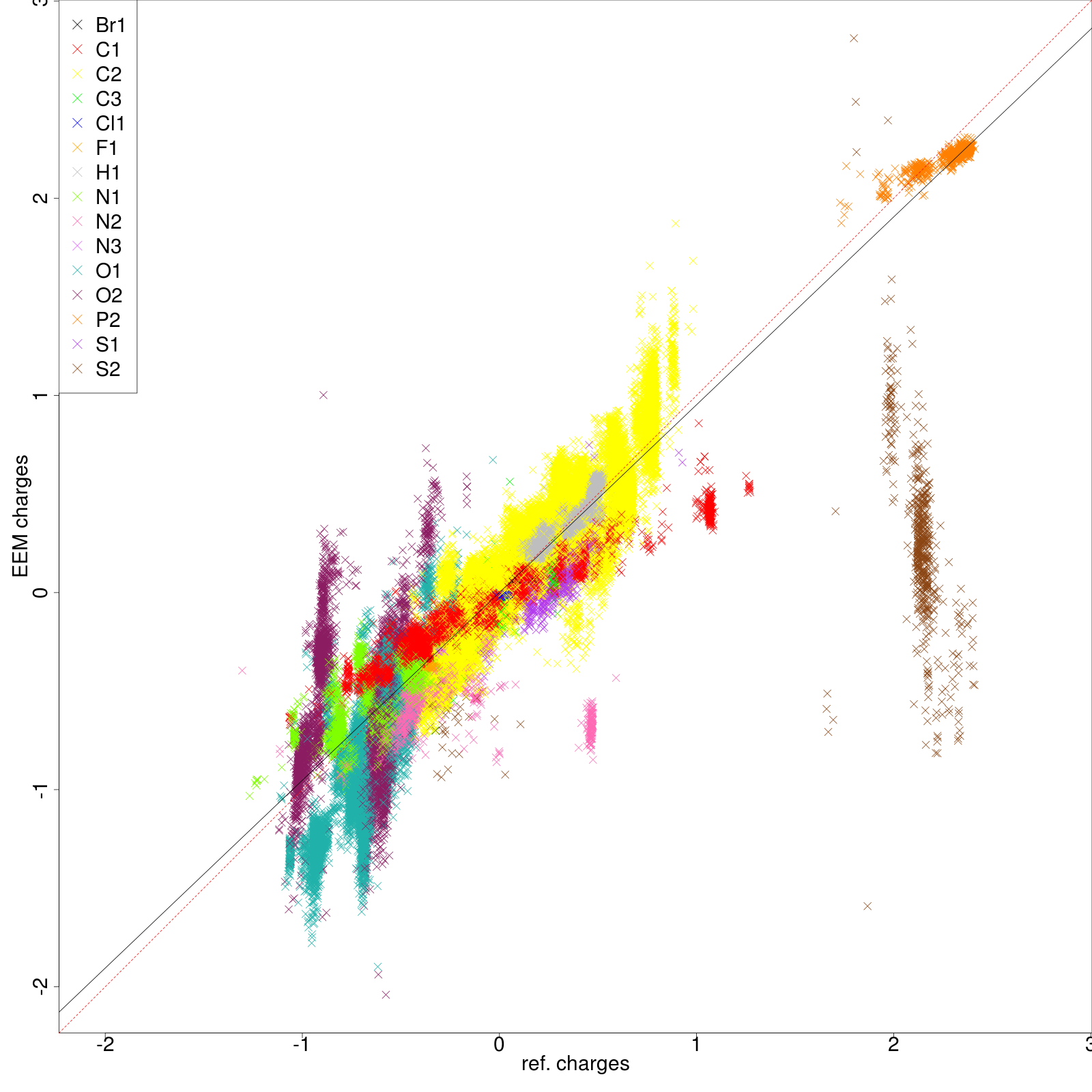
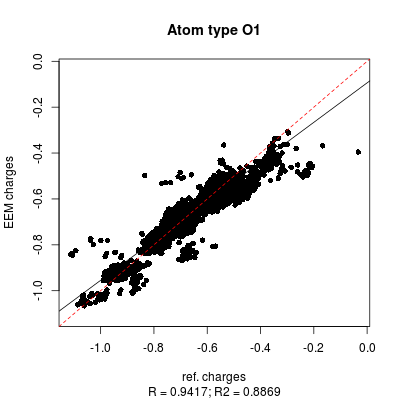
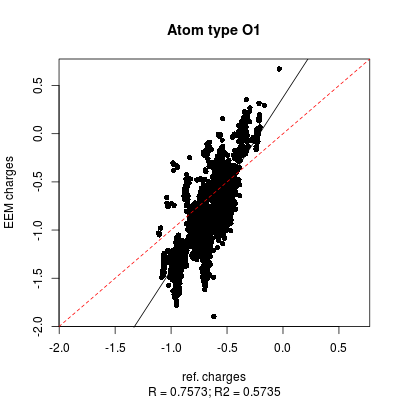
- png files displaying the charge correlation graphs for the whole set and for each atomic type (as shown in figure 6 and figure 7 respectively)
- html file gathering together all the previous information in an interactive and more easily readable report page (see figure 8 and figure 9 or click here for a more comprehensive view)
|
|
| Figure 6: All atom correlation graph between QM charges (x axis) and EEM charges (y axis). | |
|
|
| Figure 7: | |
|
|
| Figure 8: | |