NEEMP:Examples: Difference between revisions
No edit summary |
|||
| Line 6: | Line 6: | ||
=Example 1 - Training set information= | =Example 1 - Training set information= | ||
;./neemp -m info --atom-types-by ElemBond --sdf-file examples/set01.sdf | ;./neemp -m info --atom-types-by ElemBond --sdf-file examples/set01.sdf | ||
| Line 13: | Line 14: | ||
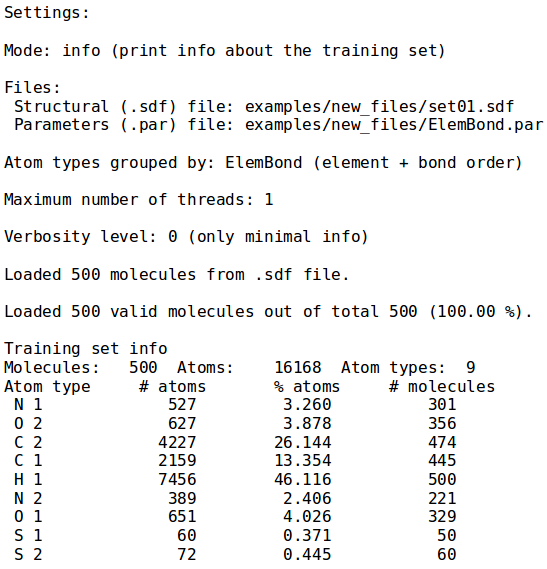
[[File:info_mode.png | thumb | 400px | center | '''''Figure 10:''''' Information about structures set ''examples/set01.sdf''. Execution time and loaded molecules number are also printed.]] | [[File:info_mode.png | thumb | 400px | center | '''''Figure 10:''''' Information about structures set ''examples/set01.sdf''. Execution time and loaded molecules number are also printed.]] | ||
=Example 2 | =Example 2 - Calculation mode= | ||
;./neemp -m charges --sdf-file examples/set01.sdf --par-file examples/ElemBond.par --chg-out-file eem_charges --max-threads 8 | ;./neemp -m charges --sdf-file examples/set01.sdf --par-file examples/ElemBond.par --chg-out-file eem_charges --max-threads 8 | ||
:Compute EEM charges and store them into the file ''eem_charges''. Use up to 8 threads for computation. | :Compute EEM charges and store them into the file ''eem_charges''. Use up to 8 threads for computation. | ||
For more information on this mode and its output, refer to [[NEEMP:Modes#Calculation mode | Calculation mode]] section. | For more information on this mode and its output, refer to [[NEEMP:Modes#Calculation mode | Calculation mode]] section. | ||
| Line 33: | Line 24: | ||
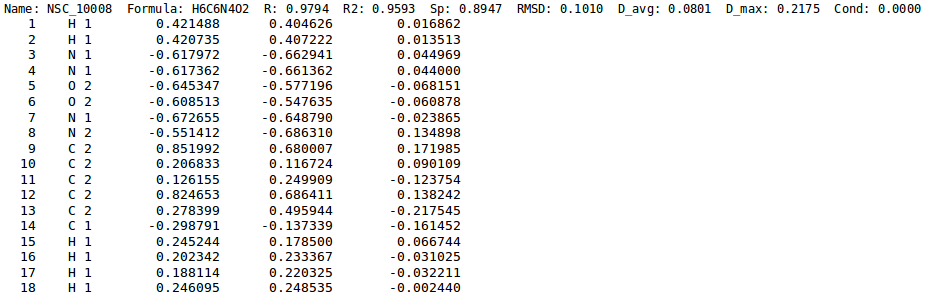
For details on ''eem_charges'' output file, see [[NEEMP:files#CHG file | CHG file]] paragraph, as the format is the same. The only different regards the third column, since in this case the '''EEM charges''' are listed in place of the '''ab-initio charges'''. | For details on ''eem_charges'' output file, see [[NEEMP:files#CHG file | CHG file]] paragraph, as the format is the same. The only different regards the third column, since in this case the '''EEM charges''' are listed in place of the '''ab-initio charges'''. | ||
=Example | =Example 3 - Parametrization mode= | ||
;./neemp -m params --sdf-file examples/set01.sdf --chg-file examples/set01.chg --par-out-file new_parameters.par --chg-stats-out-file stats | ;./neemp -m params --sdf-file examples/set01.sdf --chg-file examples/set01.chg --par-out-file new_parameters.par --chg-stats-out-file stats | ||
| Line 54: | Line 34: | ||
For additional information click [[NEEMP:Modes#Linear Regression (LR) | here]]. | For additional information click [[NEEMP:Modes#Linear Regression (LR) | here]]. | ||
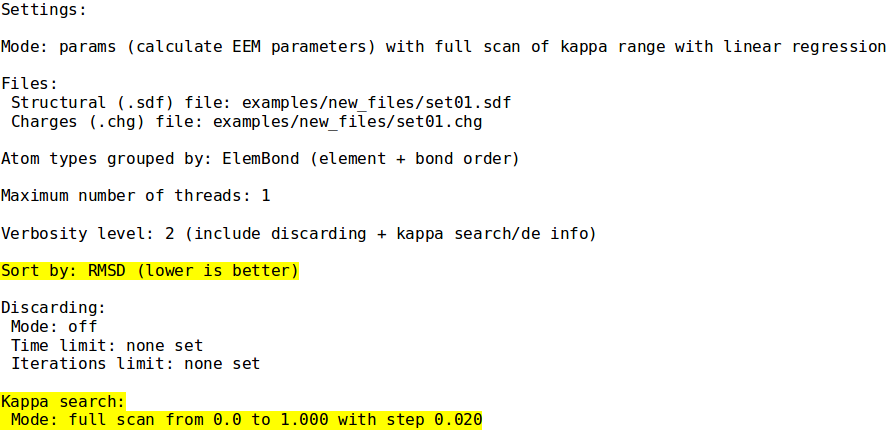
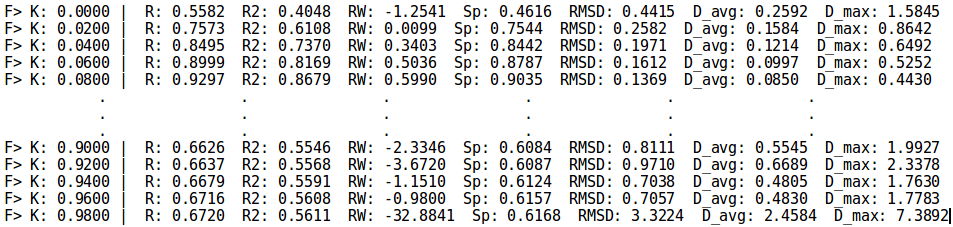
=Example | =Example 4 - Parametrization mode k search= | ||
;./neemp -m params --sdf-file examples/set01.sdf --chg-file examples/set01.chg --kappa-max 1.0 --fs-precision 0.02 --sort-by RMSD -vv | ;./neemp -m params --sdf-file examples/set01.sdf --chg-file examples/set01.chg --kappa-max 1.0 --fs-precision 0.02 --sort-by RMSD -vv | ||
| Line 71: | Line 51: | ||
|} | |} | ||
=Example | =Example 5 - Parametrization mode simple discard= | ||
;./neemp -m params --sdf-file examples/set01.sdf --chg-file examples/charges.chg --discard simple -v --check-charges --limit-iters 600 --limit-time <nowiki>00:30:00</nowiki> | ;./neemp -m params --sdf-file examples/set01.sdf --chg-file examples/charges.chg --discard simple -v --check-charges --limit-iters 600 --limit-time <nowiki>00:30:00</nowiki> | ||
| Line 80: | Line 60: | ||
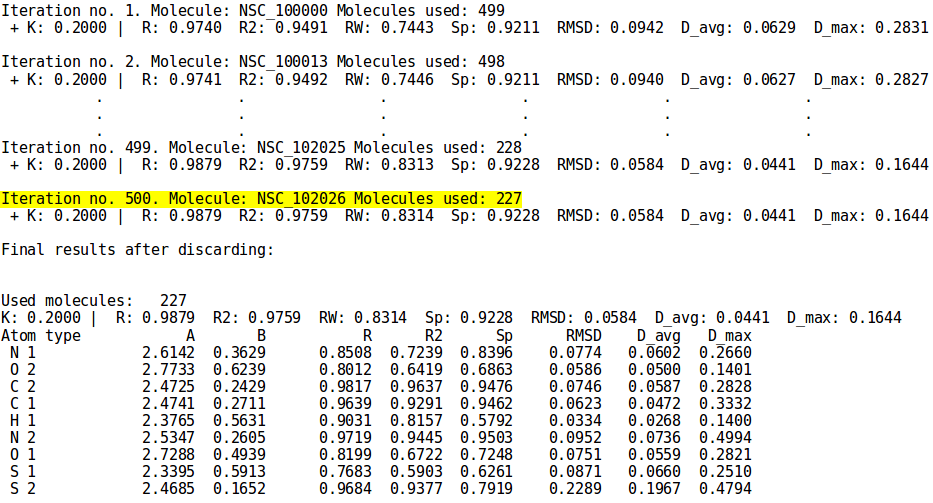
[[File:simple_discard.png | thumb | 800px | center | '''''Figure 15:''''' Abridged output for the simple discard progress. The simple discarding requires as many iterations as the number of molecules in the set, unless the iterations or time limits are reached first. In this case the training set contains only 500 structures so the iteration limit doesn't affect the discarding. The time limit still holds on the other hand. Refer to the [[NEEMP:Modes#Linear Regression (LR) | parametrization paragraph]] for details.]] | [[File:simple_discard.png | thumb | 800px | center | '''''Figure 15:''''' Abridged output for the simple discard progress. The simple discarding requires as many iterations as the number of molecules in the set, unless the iterations or time limits are reached first. In this case the training set contains only 500 structures so the iteration limit doesn't affect the discarding. The time limit still holds on the other hand. Refer to the [[NEEMP:Modes#Linear Regression (LR) | parametrization paragraph]] for details.]] | ||
=Example | =Example 6 - Parametrization mode DE-MIN= | ||
;./neemp -m params -p de --sdf-file examples/set01.sdf --chg-file examples/charges.chg --om-pop-size 50 --om-iters 500 --random-seed 1234 -vv | ;./neemp -m params -p de --sdf-file examples/set01.sdf --chg-file examples/charges.chg --om-pop-size 50 --om-iters 500 --random-seed 1234 -vv | ||
| Line 87: | Line 67: | ||
For additional information click [[NEEMP:Modes#Differential Evolution + Minimization (DE-MIN) | here]]. | For additional information click [[NEEMP:Modes#Differential Evolution + Minimization (DE-MIN) | here]]. | ||
=Example 7 - Coverage validation= | |||
;./neemp -m cover -sdf-file examples/set02.sdf --par-file examples/Element.par --atom-types-by Element | |||
:Calculate coverage of supplied EEM parameters and molecules set. | |||
<br style="clear:both" /> | |||
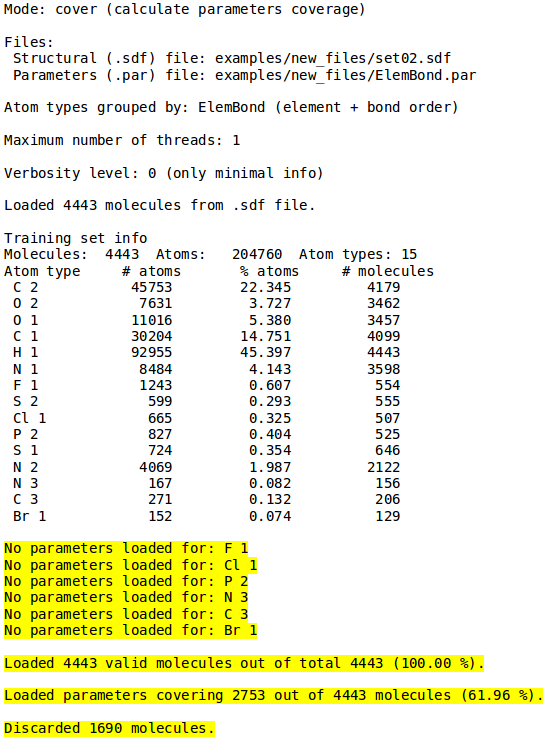
[[File:cover_mode2.png | thumb | 400px | center | '''''Figure 11:''''' Complete output for the ''Cover mode'' summarizing information about the input files, molecules set composition and coverage. For more information on this mode refer to [[NEEMP:Modes#Coverage validation | Cover validation]] section.]] | |||
=Example 8 - Quality validation= | |||
;./neemp -m cross --sdf-file examples/set02.sdf --chg-file examples/charges.chg --par-file examples/Element.par --chg-stats-out stats --atom-types-by Element 2> warns > log | |||
:Perform cross-validation of EEM parameters for atoms grouped by element only. Save standard output into the file ''log'' and warnings into the file ''warns''. Moreover outputs charge statistics for each molecule into the file ''stats''. | |||
For more information on this mode click [[NEEMP:Modes#Quality validation | here]]. | |||
<br style="clear:both" /> | |||
[[File:chg_stats.png | thumb | 900px | center | '''''Figure 12:''''' Close-up from ''stats'' file. Along with statistics for each molecule, ''ab-initio'' charges (3rd column), ''EEM'' charges (4th column) and their difference (5th column) are also printed out.]] | |||
Revision as of 06:24, 28 June 2016
This section shows several use case examples. All of them use only data from examples directory.
NB: NEEMP is case-sensitive. Please follow carefully this section for insight on the correct syntax. Additional information can be found here.
Example 1 - Training set information
- ./neemp -m info --atom-types-by ElemBond --sdf-file examples/set01.sdf
- Prints information about training set set01.sdf, provided as argument of the
--sdf-fileoption, grouping atoms according to chemical element and bond order. To override this behaviour and group atoms only by element, call NEEMP with--atom-types-by Elementoption.
Example 2 - Calculation mode
- ./neemp -m charges --sdf-file examples/set01.sdf --par-file examples/ElemBond.par --chg-out-file eem_charges --max-threads 8
- Compute EEM charges and store them into the file eem_charges. Use up to 8 threads for computation.
For more information on this mode and its output, refer to Calculation mode section.
For details on eem_charges output file, see CHG file paragraph, as the format is the same. The only different regards the third column, since in this case the EEM charges are listed in place of the ab-initio charges.
Example 3 - Parametrization mode
- ./neemp -m params --sdf-file examples/set01.sdf --chg-file examples/set01.chg --par-out-file new_parameters.par --chg-stats-out-file stats
- Performs EEM parametrization, saves parameters into the file new_parameters.par, outputs charge statistics for each molecule into the file stats. No discarding is used.
NB: the parameters set file new_parameters.par presents the same identical format and layout as described here.
For additional information click here.
Example 4 - Parametrization mode k search
- ./neemp -m params --sdf-file examples/set01.sdf --chg-file examples/set01.chg --kappa-max 1.0 --fs-precision 0.02 --sort-by RMSD -vv
- Similar to previous example, use custom range for k. Select best parameters according to RMSD. Print k search progress.
Reference to parametrization paragraph and options list if necessary.
Example 5 - Parametrization mode simple discard
- ./neemp -m params --sdf-file examples/set01.sdf --chg-file examples/charges.chg --discard simple -v --check-charges --limit-iters 600 --limit-time 00:30:00
- Perform simple discarding and print its progress. Issue warning about molecules with abnormal values of statistical descriptors. The duration of the discarding procedure can be at most 600 iterations or 30 minutes (time format HH:MM:SS), whichever is reached first.
Example 6 - Parametrization mode DE-MIN
- ./neemp -m params -p de --sdf-file examples/set01.sdf --chg-file examples/charges.chg --om-pop-size 50 --om-iters 500 --random-seed 1234 -vv
- Compute parameters for the given molecules in file set01.sdf and ab-initio charges in charges.chg. The chosen optimization method: differential evolution + minimization will act on a parameter vector population consisting of 50 units. The optimization procedure will be carried out for at most 500 iterations. A user-defined seed is used, as well as an high level of verbosity.
For additional information click here.
Example 7 - Coverage validation
- ./neemp -m cover -sdf-file examples/set02.sdf --par-file examples/Element.par --atom-types-by Element
- Calculate coverage of supplied EEM parameters and molecules set.
Example 8 - Quality validation
- ./neemp -m cross --sdf-file examples/set02.sdf --chg-file examples/charges.chg --par-file examples/Element.par --chg-stats-out stats --atom-types-by Element 2> warns > log
- Perform cross-validation of EEM parameters for atoms grouped by element only. Save standard output into the file log and warnings into the file warns. Moreover outputs charge statistics for each molecule into the file stats.
For more information on this mode click here.