NEEMP:Modes: Difference between revisions
| Line 41: | Line 41: | ||
<br style="clear:both" /> | <br style="clear:both" /> | ||
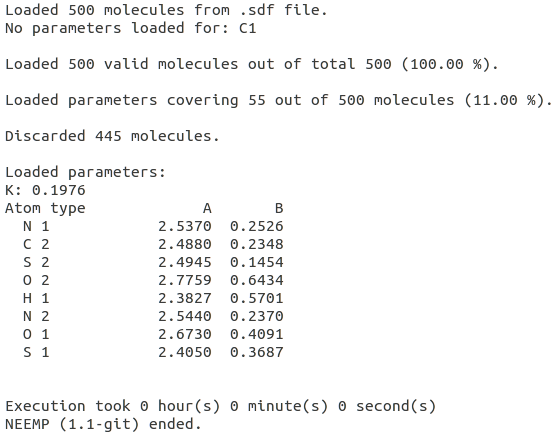
[[File:cross_mode.png | thumb | | [[File:cross_mode.png | thumb | 600px | center | '''''Figure 4:''''' The percentage of covered molecules in the set is extremely low because of the absence of parameters for C1. All the molecules containing this atom types are automatically discarded.]] | ||
Revision as of 12:26, 30 May 2016
NEEMP features several functionalities, each of those can be selected by the option --mode or -m combined with one of the following keywords:
info : display information about the training set
cover : display how many molecules are covered by the given parameters set
charges : calculate EEM charges
cross : perform cross-validation
params : calculate EEM parameters (due to its complexity and relevance this mode is treated on a separate section)
Info (-m info)
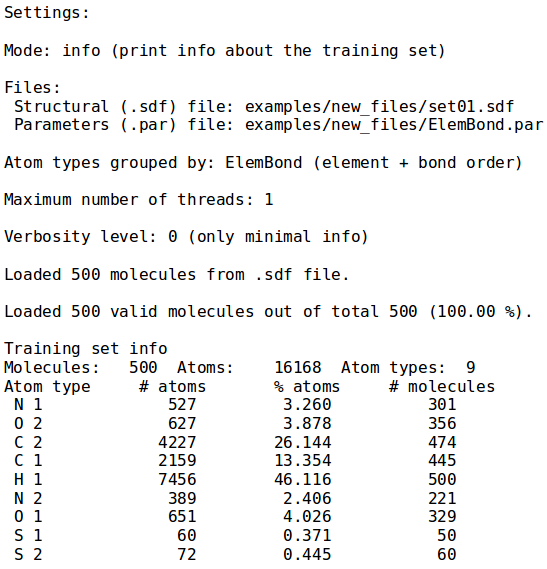
The first mode shows information about the training set provided as argument to the --sdf-file option. Specifically, number of molecules, the total number of atoms and number of atoms for each type are printed; the latter being defined by default as the chemical element and the maximum bond order. To override this behaviour in all NEEMP's modes and group the atoms only by chemical element, use the option --atom-types-by Element. Figure 1 displays the output for set01 in the examples directory, with default grouping, specified by ElemBond keyword (for details on the syntax, see the examples or option list paragraph).
Cover (-m cover)
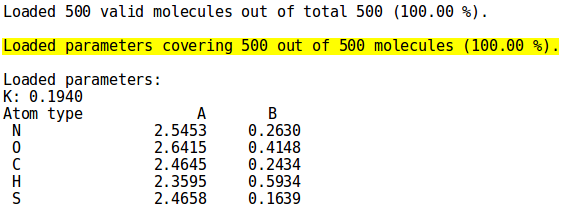
To see how many molecules from the set are covered by the provided parameters, simply pass NEEMP those two files (options --sdf-file and --par-file). Figure 2 shows a close-up from the cover mode output for set02.sdf and the parameters set Element.par, both present in the examples directory (for details on the syntax and the complete output, see the examples or option list paragraph)
Charges (-m charges)
To compute EEM charges with NEEMP, it is necessary to provide the SDF file with molecules for which the charges will be computed (option --sdf-file) and the PAR file with EEM parameters (option --par-file). EEM charges will be written into the file specified by --chg-out-file option.
NEEMP checks if the matrix is well-conditioned (note that this won't apply for GNU version) by estimating its condition number. If its reciprocal is lower than WARN_MIN_RCOND defined in config.h, warning about possible bad charges for a particular molecule is issued.
The user should keep in mind that, because of the EEM method itself, the EEM charges cannot be computed for elements or element types for which the parameters are missing. This situation is well depicted by Figure 3, where a modified version of the parameters set examples/ElemBond.par lacking the parameters for C1 (carbon presenting only single bonds), has been employed to compute the EEM charges for the set examples/set01.sdf (for details on the syntax, see the examples or option list section).
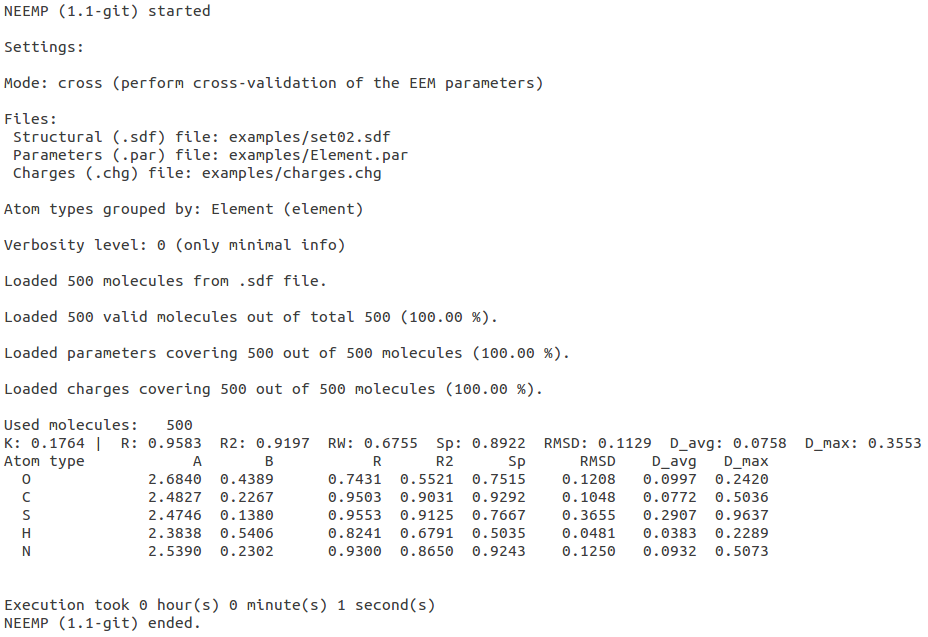
Cross-validation (-m cross)
To assess the quality of the EEM parameters on a different set than one they were based on, NEEMP's cross mode can be used.
Required input files are: EEM parameters (passed as argument of option --par-file) and a new set of molecular structures (argument of --sdf-file) for which ab-initio charges have been computed (argument of --chg-file).