NEEMP:files: Difference between revisions
No edit summary |
|||
| Line 4: | Line 4: | ||
=SDF file= | =SDF file= | ||
[[File:sdf_file.png | thumb | 500px | left| ''P. aeruginosa '' PA-IIL Lectin (1gzt) sugar binding site identified with '''PatternQuery''']] | |||
'''SDF''' file contains MOL records for each molecule separated by line consisting only of four dollar signs, i.e. $$$$. Each MOL record can be either in '''V2000''' or '''V3000''' version. The latter one is required for molecules with more than 999 atoms or bonds. For further reference on MOL format, see specification: [http://c4.cabrillo.edu/404/ctfile.pdf CTFile formats]. | '''SDF''' file contains MOL records for each molecule separated by line consisting only of four dollar signs, i.e. $$$$. Each MOL record can be either in '''V2000''' or '''V3000''' version. The latter one is required for molecules with more than 999 atoms or bonds. For further reference on MOL format, see specification: [http://c4.cabrillo.edu/404/ctfile.pdf CTFile formats]. | ||
Revision as of 11:07, 29 May 2016
NEEMP uses 3 types of input files. SDF file contains structural information about the molecules, i.e. positions of atoms and bonding information; CHG file contains previously computed ab-initio charges; and PAR file stores the list of NEEMP's EEM parameters.
The following sections illustrate the details of each file type. Examples of these input files can also be found in the examples directory.
SDF file
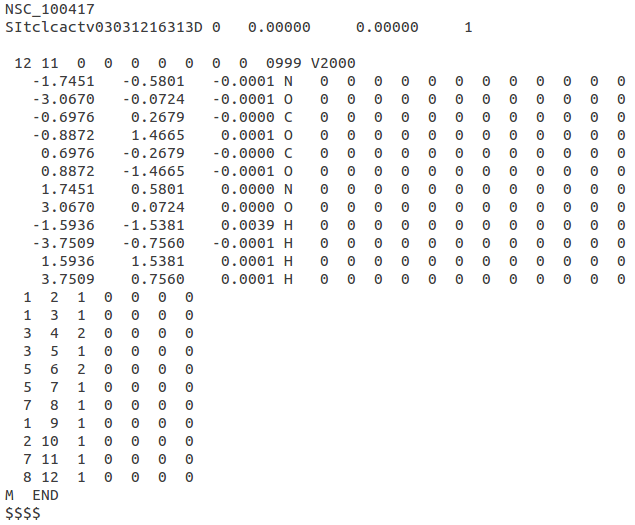
SDF file contains MOL records for each molecule separated by line consisting only of four dollar signs, i.e. $$$$. Each MOL record can be either in V2000 or V3000 version. The latter one is required for molecules with more than 999 atoms or bonds. For further reference on MOL format, see specification: CTFile formats.
For convenience NEEMP has a support for reading SDF files which are compressed using gzip method. This might be useful when working with large databases of molecules.