ChargeCalculator:Case studies: Difference between revisions
| Line 16: | Line 16: | ||
=Protegrin-1 – atomic charges and activity of antimicrobial peptides= | =Protegrin-1 – atomic charges and activity of antimicrobial peptides= | ||
[[File:Protegrin.png|thumb|right|300px| | [[File:Protegrin.png|thumb|right|300px| Residue centroids represented by spheres colored by residue charges. Residues ARG4 and ARG9 are significantly less positive (lighter blue) than the rest of the ARG residues (darker blue), indicating that mutations at these positions may be less effective.]] | ||
Protegrins are a family of antimicrobial peptides active against a wide range of pathogens. Protegrin-1 (PG1) has been intensely studied for its potential in treating infections caused by antibiotic resistant bacteria. PG1 shows activity against several pathogens, but also toxicity against the host. Useful mutations are those which maintain the antimicrobial activity, and at the same time reduce toxicity. | Protegrins are a family of antimicrobial peptides active against a wide range of pathogens. Protegrin-1 (PG1) has been intensely studied for its potential in treating infections caused by antibiotic resistant bacteria. PG1 shows activity against several pathogens, but also toxicity against the host. Useful mutations are those which maintain the antimicrobial activity, and at the same time reduce toxicity. | ||
| Line 25: | Line 25: | ||
You can try out this particular computation setup [http://webchem.ncbr.muni.cz/Platform/ChargeCalculator/Result/dc41612f-b3c0-4207-97a6-b70598e90d88?example=Protegrin here], and make sure to use the interactive guides for additional explanations. The total molecular charge was +7, owing to the many ARG residues. The EEM parameter set used was EX-NPA_6-31Gd_gas, but it was necessary to add to this set EEM parameters for deuterium (D), because this element was present in the input file. The EEM parameters for D were identical to the parameters for H. The EEM parameter set covers NPA atomic charges at the HF/6-31G* level of theory. These charges are expected to support qualitative chemical concepts like induction and conjugation. The rest of the ACC default settings were kept. | You can try out this particular computation setup [http://webchem.ncbr.muni.cz/Platform/ChargeCalculator/Result/dc41612f-b3c0-4207-97a6-b70598e90d88?example=Protegrin here], and make sure to use the interactive guides for additional explanations. The total molecular charge was +7, owing to the many ARG residues. The EEM parameter set used was EX-NPA_6-31Gd_gas, but it was necessary to add to this set EEM parameters for deuterium (D), because this element was present in the input file. The EEM parameters for D were identical to the parameters for H. The EEM parameter set covers NPA atomic charges at the HF/6-31G* level of theory. These charges are expected to support qualitative chemical concepts like induction and conjugation. The rest of the ACC default settings were kept. | ||
<br style="clear:both" /> | |||
=Proteasome= | =Proteasome= | ||
Revision as of 18:21, 14 June 2015
We present a few interesting examples of use cases for ACC.
Paracetamol – atomic charges and chemical reactivity in small drug like molecules
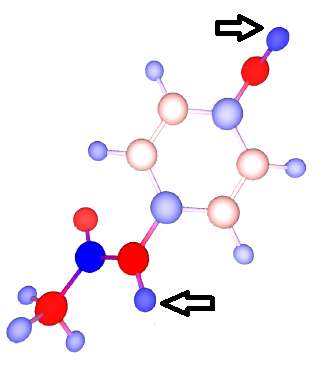
N-acetyl-p-aminophenol, commonly known as paracetamol, is a widely used analgesic and antipyretic. Its mechanism of action is believed to be the inhibition of the protein cyclooxygenase 2, regulating the production of pro-inflammatory compounds. The metabolic break down of paracetamol has been the subject of intense study, since it holds the key to both its therapeutic action and toxicity.
We calculated atomic charges in paracetamol using ACC. The geometry of the paracetamol molecule corresponded to the ideal coordinates for the entry TYL from the wwPDB Chemical Compounds Database (wwPDB CCD: TYL). The default ACC settings were used. The computation took less than 1s, and the complete results are available on the ACC web page.
A quick analysis reveals that the phenolic H (position HO4) is the most acidic proton (highest positive charge) in the molecule, suggesting a faster and easier dissociation of this O-H bond. Indeed up to 90% of metabolic degradation happens at position HO4 (glucunronidationration, sulphonation). Additionally, up to 15% of metabolic degradation involves dehydrogenation oxidation at the phenolic (HO4) and amidic positions (HN), the two most positive H in the paracetamol molecule. While paracetamol is a very small molecule with few polar sites, the same principle can be applied in reasoning out highly reactive sites in more complex molecules.
You can try out this particular computation setup here, and make sure to use the interactive guides for additional explanations. We have used the default ACC settings, meaning that the total charge was 0, the EEM parameter set covers NPA atomic charges at the B3LYP/6-31G* level of theory. These charges are expected to support qualitative chemical concepts like induction and conjugation. In general, it is a good idea to try several charge definitions. One or more of these might prove useful in further modeling studies. If you plan to run molecular dynamics, it is good to use charges which are compatible with the particular force field you plan to use. For instance, if the force field is known to use atomic charges based on electrostatic potential mapping, pick at least one EEM parameter set developed with this charge definition.
Protegrin-1 – atomic charges and activity of antimicrobial peptides
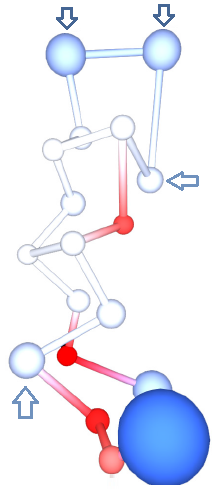
Protegrins are a family of antimicrobial peptides active against a wide range of pathogens. Protegrin-1 (PG1) has been intensely studied for its potential in treating infections caused by antibiotic resistant bacteria. PG1 shows activity against several pathogens, but also toxicity against the host. Useful mutations are those which maintain the antimicrobial activity, and at the same time reduce toxicity.
We calculated atomic charges in PG1 using ACC. The geometry of the PG1 molecule corresponded to the lowest energy NMR model in the entry 1PG1 (PDB: 1PG1) from the Protein Data Bank. The computation took less than 1s, and the complete results are available on the ACC web page.
The calculation produced one set of atomic charges. PG1 contains 18 residues, and rather than analyzing atomic charges, we analyzed the residue charges, which are also reported by ACC (Figure 4B). PG1 is special because of its high positive charge. It contains 6 ARG residues. However, not all have the same charge. In particular, ARG at positions 4 and 9 have the least positive charge (around +0.5e), whereas the rest have much higher positive charge (over +0.8e). Such results suggest that mutations of ARG into a neutral residue at positions 4 or 9 would have a lower effect than mutations at positions 1, 10, 11 or 18. This seems to be indeed the case, as it was found that the mutation of ARG 4 alters the antimicrobial activity against C. albicans significantly less than the mutation of ARG 10. Such biologically relevant insight can be gained by analyzing the residue charges on a single structure of PG1.
You can try out this particular computation setup here, and make sure to use the interactive guides for additional explanations. The total molecular charge was +7, owing to the many ARG residues. The EEM parameter set used was EX-NPA_6-31Gd_gas, but it was necessary to add to this set EEM parameters for deuterium (D), because this element was present in the input file. The EEM parameters for D were identical to the parameters for H. The EEM parameter set covers NPA atomic charges at the HF/6-31G* level of theory. These charges are expected to support qualitative chemical concepts like induction and conjugation. The rest of the ACC default settings were kept.
Proteasome
The 20S subunit is the catalytic machinery of proteasomes, whose main function is to break down unneeded or damaged proteins. The 20S proteasome is a cylindrical nanoparticle with a wide hollow core, where protein degradation takes place at multiple catalytic sites. The proteolysis mechanism is enabled by a threonine-dependent nucleophilic attack, which may depend on associated water molecules for deprotonation of the reactive threonine hydroxyl. It is possible to study the influence of water molecules on the charge distribution of the catalytic threonine before moving on to more complex modeling studies of the catalytic reaction itself.
There are 7 beta catalytic units in the 20s proteasome, each made up of two chains (H-N,V-Z,1,2). The catalytic THR residues occupy the N-terminal position of each chain, so its residue serial number will be 1, and we expect it to be positively charged. In eukariotes, only 4 of these beta subunits contain the expected catalytic THR residues (chains H,I,L,N,V,W,Z,2).
For studying the potential influence of water on the catalytic THR, we have calculated atomic charges for the 20S proteasome of cattle (PDB ID 1IRU) using an EEM parameter set specifically developed for proteins, since the 20S proteasome is actually a large complex of multiple proteins. Try out this particular computation setup here, and make sure to use the interactive guides for additional explanations.
The results of the calculation are available here.
The presence of water molecules generally makes the catalytic THR residues less positive by up to 0.08e. This effect is favorable to the catalytic activity, as the THR hydrogen is partially drawn to water, enabling the reactive hydroxyl to initiate the nucleophilic attack. Maximum effect is seen where water forms an H-bond with the THR hydroxyl (e.g., THR 1 from chain W). A smaller effect is seen where the water is not in optimal position to form an H bond (e.g., THR 1 from chain V). The only exception is the catalytic THR of chain L, where the presence of water actually seems to make the THR more positive by over 0.2e. This is caused by the presence of not one, but three water molecules in close proximity of THR, and is probably related to the different substrate specificity of this site.
Note that the input file also contains Mg ions, which were probably used to stabilize the 20S proteasome in the original experiment that produced the structure with PDB ID 1iru. The influence of Mg ions on the catalytic THR can also be studied in the same straightforward fashion as above. Simply remove the ions from the input file, run ACC with the same setup, and then compare the charge on the catalytic THR residues between the two computations (with and without Mg ions).
Return to the Table of contents.