PatternQuery:Introduction: Difference between revisions
| Line 28: | Line 28: | ||
=Where should I start?= | =Where should I start?= | ||
Before you write your first query, it is many times useful to go through our manual, on [[MotiveQuery:How to build a query | how to write a query]] or take a look into the [[MotiveQuery:Language Reference | language reference]]. If you do not feel like reading anything, you can try and browse one of our ready-to-use samples or examples, which are ready at '''MotiveQuery''' interface for your convenience. If neither of them helps, ''Ask us'' a question via a simple | Before you write your first query, it is many times useful to go through our manual, on [[MotiveQuery:How to build a query | how to write a query]] or take a look into the [[MotiveQuery:Language Reference | language reference]]. If you do not feel like reading anything, you can try and browse one of our ready-to-use samples or examples, which are ready at '''MotiveQuery''' interface for your convenience. If neither of them helps, ''Ask us'' a question via a simple [http://webchem.ncbr.muni.cz/Platform/MotiveQuery/Index#support-tab| user interface]. We will do our best to get back to you as soon as possible. We are always happy to answer your questions related to the service, or even to help you constructing your queries for your own research. | ||
'''Enjoy working with MotiveQuery!''' | '''Enjoy working with MotiveQuery!''' | ||
Revision as of 20:20, 28 December 2014
All the life sciences, namely structural biology, bioinformatics or computational chemistry benefits from the rapid increase of data publicly available. The amount and quality of biomacromolecular sequence and structural data were increasingly growing in the past decade and it seems that the trend isn't to slow down any time soon. This stresses enormous emphasis on the availability of tools for processing and interpreting these data, since deciphering previously unknown features and dependencies sheds more light on a variety of biological processes. Comparative analysis of proteins or its parts helps to uncover their function. Thorough analyses of protein binding schemes inevitably leads for example to the preparation of more stable and versatile catalysis, or helps to find suitable inhibitors of different biological pathways.
What is MotiveQuery?
MotiveQuery Service is an interface to the MotiveQuery language simple, yet universal and versatile chemical language. The language allows you to perform a variety of searches - from simple identification of ions or ligands and their binding sites, through detection of specific binding motifs such as zinc-fingers, to complex pharmacophores modulating biological activity. These searches are performed over the largest database of biomacromolecular structures - Protein Data Bank.
How it can help me?
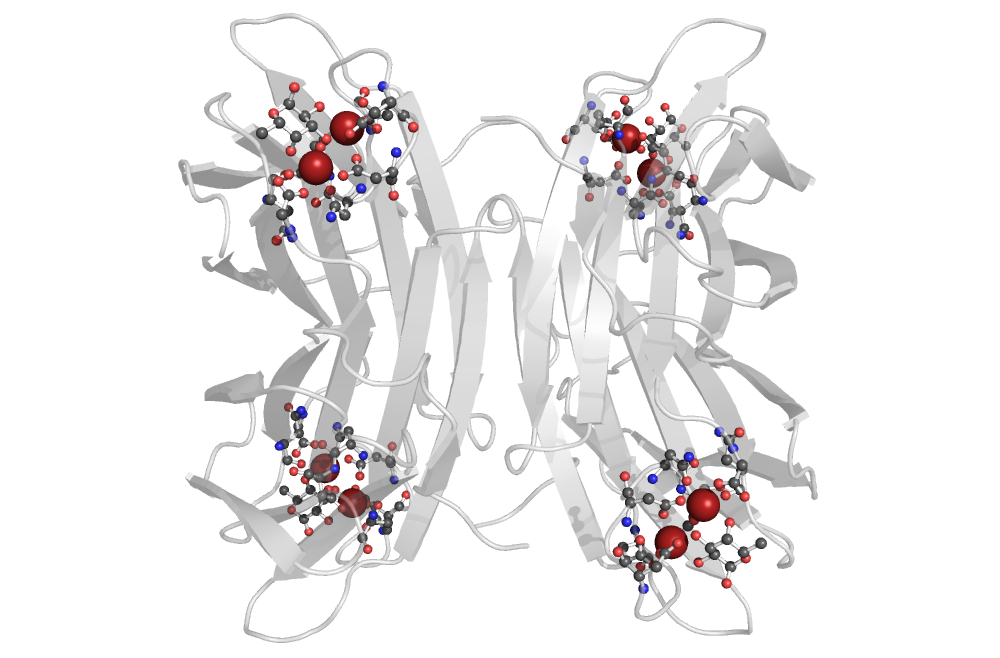
MotiveQuery is tailored to describe biomacromolecular structural fragments based on the nature and the relationship between atoms, residues, and other structural elements, both at the sequence and 3D level. This unique design allows the user to simultaneously operate and combine several levels of resolution to identify fragments of biomacromolecular complexes including ligands, water molecules, and ions. Such knowledge have a variety of imminent applications, for example: in a structural and functional assignment of uncharacterized or newly determined proteins, but also represents a key point in rational design and engineering of novel functional sites and comparative protein structural analyses.
How can I use it?
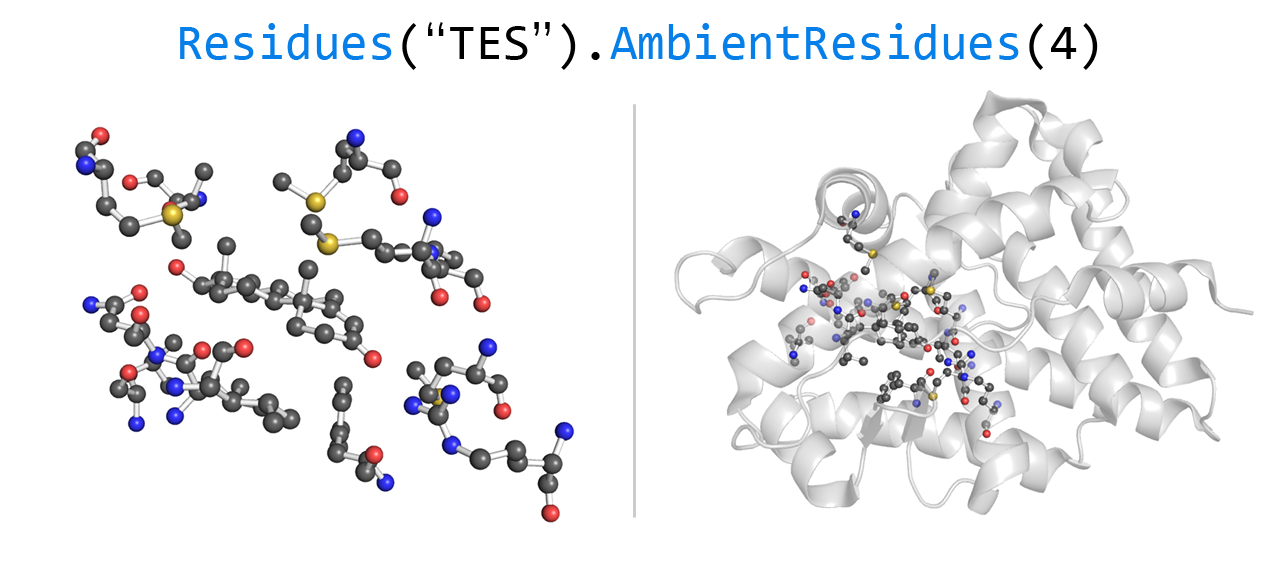
The service is available in 3 different versions:
- MotiveQuery Explorer for querying smaller data sets, or user uploaded data. However its main advantage lies in perfecting queries before one performs PDB-wide searches.
- MotiveQuery Service - a web HTML/JavaScript interface for the MotiveQuery Service. PDB database or its subset based on a variety of different properties, such as Enzymatic number, Organism of Origin or Resolution.
- Command-line application an offline version of MotiveQuery aimed for querying in house copies of different structural databases.
For further information on how to control the application please take a look at the relevant wiki pages, or directly use the application. Whenever you are unsure on how to carry on, use one of the many guides (reachable by ? button) which are there for your convenience.
Where should I start?
Before you write your first query, it is many times useful to go through our manual, on how to write a query or take a look into the language reference. If you do not feel like reading anything, you can try and browse one of our ready-to-use samples or examples, which are ready at MotiveQuery interface for your convenience. If neither of them helps, Ask us a question via a simple user interface. We will do our best to get back to you as soon as possible. We are always happy to answer your questions related to the service, or even to help you constructing your queries for your own research.
Enjoy working with MotiveQuery!