PatternQuery:Specifics Page: Difference between revisions
Created page with "The '''MotiveQuery''' specifics page provides detailed information about detected fragments and their parent structures. All the results can be downloaded and further inspecte..." |
No edit summary |
||
| Line 8: | Line 8: | ||
| [[File:MotiveQuery-Service-Results2.png| 650px]] | | [[File:MotiveQuery-Service-Results2.png| 650px]] | ||
|- | |- | ||
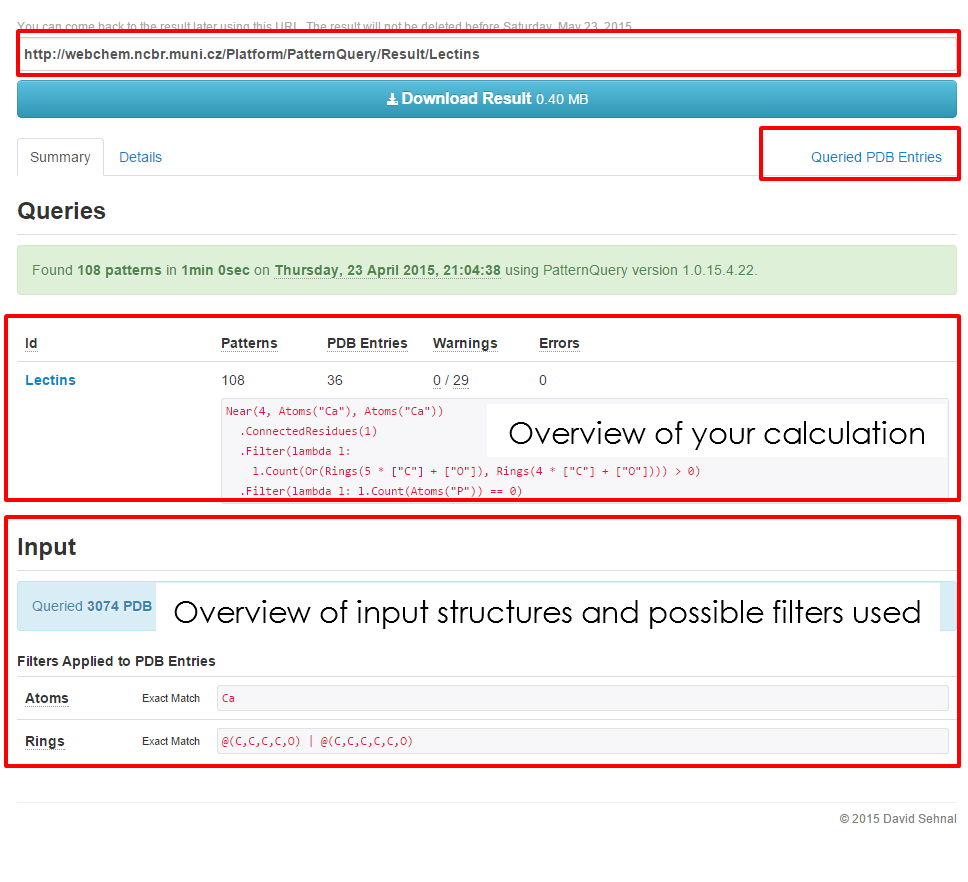
| The Summary of the '''Specifics page''' is divided into the several panels. At first bookmark the web address, to come back later. Additionally, you can download all the provided results for their later inspection. The summary results of the calculation, | | The Summary of the '''Specifics page''' is divided into the several panels. At first bookmark the web address, to come back later. Additionally, you can download all the provided results for their later inspection. The summary results of the calculation, as well as the calculation setup are highlighted below. Click '''Query PDB Entry Details''' for inspecting further details of all of the input PDB structures. | ||
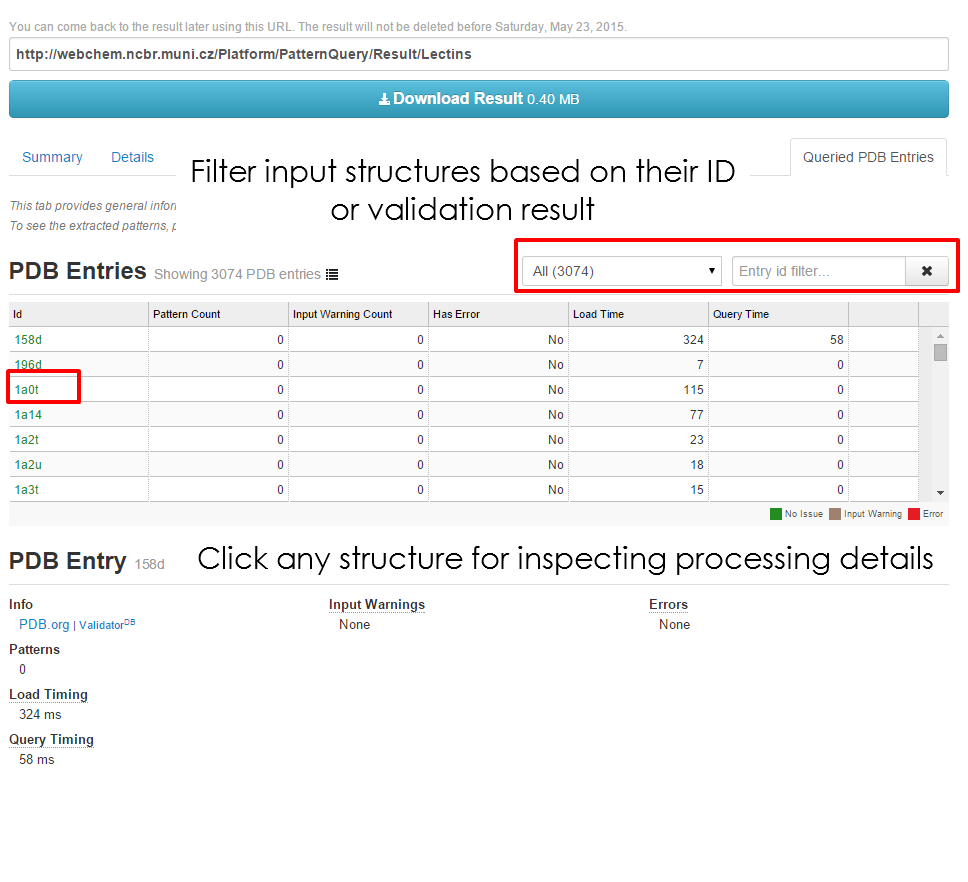
| Filter the '''Structure table''' by a structure validation | | Filter the '''Structure table''' by a structure validation result (no issues, missing atoms, chirality issues) as well as the PDB id. Click the structure id for inspecting greater details. | ||
|- | |- | ||
| [[File:MotiveQuery-Service-Results3.png| 650px]] | | [[File:MotiveQuery-Service-Results3.png| 650px]] | ||
| Line 17: | Line 17: | ||
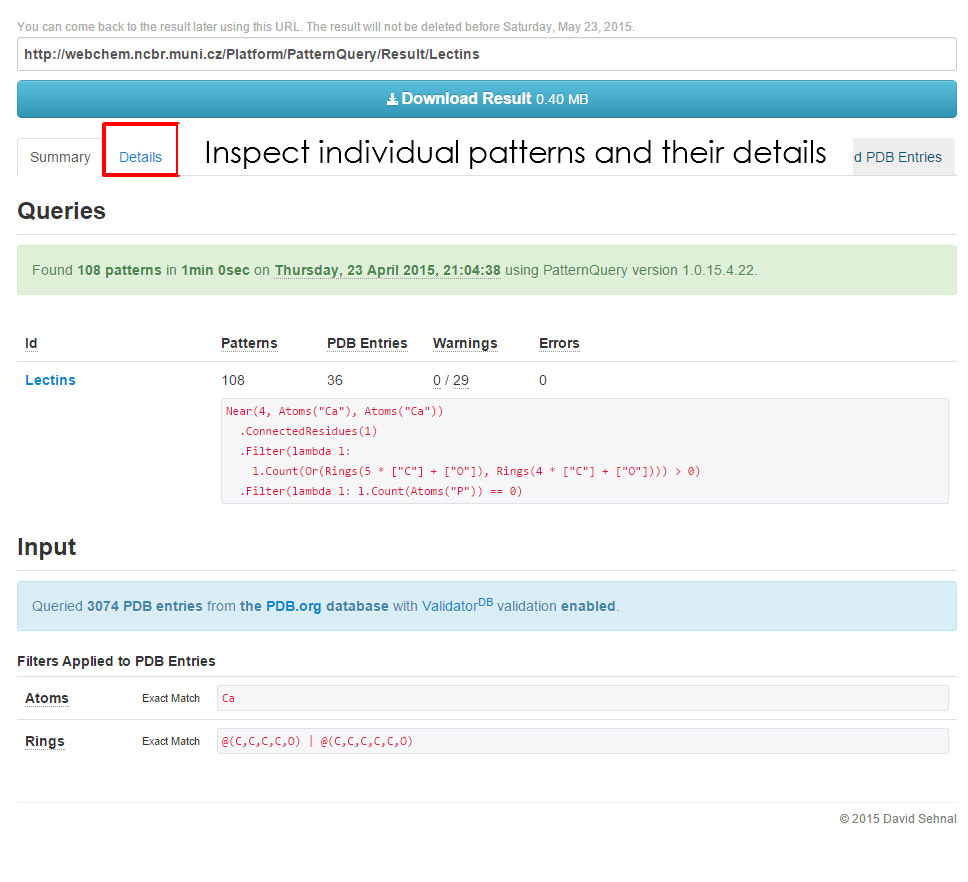
==Fragment Details== | ==Fragment Details== | ||
'''Fragment details''' tab | '''Fragment details''' tab provides valuable information about the composition and origin of the fragment. The structure validation report is provided for your convenience for all ligands and non-standard residues larger 6 atoms as well. | ||
{| class="wikitable" border="1" style="margin: 1em 1em 1em 1em;" width="650px" | {| class="wikitable" border="1" style="margin: 1em 1em 1em 1em;" width="650px" | ||
| Line 24: | Line 24: | ||
| [[File:MotiveQuery-Fragments2.png| 650px]] | | [[File:MotiveQuery-Fragments2.png| 650px]] | ||
|- | |- | ||
| Select one of the queries for inspecting individual fragments. Browse the '''Fragment table''' and inspect details of particular fragment | | Select one of the queries for inspecting individual fragments. Browse the '''Fragment table''' and inspect details of particular fragment as well as its visualization. | ||
| Filter the '''Fragment table''' by a structure validation | | Filter the '''Fragment table''' by a structure validation result (no issues, missing atoms, chirality issues) as well as the fragment Id. Filtering by a variety of different properties such as the ''Year of publication'', ''Resolution'' or ''Enzymatic number'' also works. Click the structure id for inspecting greater details. | ||
|- | |- | ||
| [[File:MotiveQuery-Fragments3.png| 650px]] | | [[File:MotiveQuery-Fragments3.png| 650px]] | ||
| Line 47: | Line 47: | ||
| [[File:MotiveQuery-Fragments7.png| 650px]] | | [[File:MotiveQuery-Fragments7.png| 650px]] | ||
|- | |- | ||
| Filter the '''Fragment table''' by a structure validation | | Filter the '''Fragment table''' by a structure validation result (no issues, missing atoms, chirality issues) as well as the fragment id. Click the structure id for inspecting greater details. | ||
| Inspect additional details of the PDB structures in different resources, such as PDB.org database or Validator<sup>DB</sup> storing their ligand validation information. | | Inspect additional details of the PDB structures in different resources, such as PDB.org database or Validator<sup>DB</sup> storing their ligand validation information. | ||
|- | |- | ||
|} | |} | ||
Revision as of 15:59, 20 April 2015
The MotiveQuery specifics page provides detailed information about detected fragments and their parent structures. All the results can be downloaded and further inspected by a different means. You can also come back later and check out the results with the web address provided.
Calculation Summary
Fragment Details
Fragment details tab provides valuable information about the composition and origin of the fragment. The structure validation report is provided for your convenience for all ligands and non-standard residues larger 6 atoms as well.
| File:MotiveQuery-Fragments1.png | File:MotiveQuery-Fragments2.png |
| Select one of the queries for inspecting individual fragments. Browse the Fragment table and inspect details of particular fragment as well as its visualization. | Filter the Fragment table by a structure validation result (no issues, missing atoms, chirality issues) as well as the fragment Id. Filtering by a variety of different properties such as the Year of publication, Resolution or Enzymatic number also works. Click the structure id for inspecting greater details. |
| File:MotiveQuery-Fragments3.png | |
| After clicking fragment ID, all the additional informations are immediately visualized together with the fragment view. |
Structure Details
Structure details tab provides a valuable information about fragment's parent structures. For example you can immediately access just fragments originating from a particular structure or structures. Filtering based on a variety of properties is available as well.
| File:MotiveQuery-Fragments4.png | File:MotiveQuery-Fragments5.png |
| Click on the By PDB Entry tab for inspecting PDB entry-wide results. | The page is divided into a fully customizable table (by structure properties) and details for an individual structure. |
| File:MotiveQuery-Fragments6.png | File:MotiveQuery-Fragments7.png |
| Filter the Fragment table by a structure validation result (no issues, missing atoms, chirality issues) as well as the fragment id. Click the structure id for inspecting greater details. | Inspect additional details of the PDB structures in different resources, such as PDB.org database or ValidatorDB storing their ligand validation information. |