File:VDB manual figures2.png: Difference between revisions
Lukas uploaded a new version of "File:VDB manual figures2.png" |
Lukas uploaded a new version of "File:VDB manual figures2.png" |
||
| (9 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
A) Overview of validation process. For each PDB entry, the relevant residues are detected based on their name (3-letter code) and number of atoms. Then, for each validated residue, the corresponding model is retrieved from wwPDB CCD, and each motif of this residue is validated against the model. ValidatorDB is then built as the collection of validation results for all motifs of all residues in all PDB entries. | A) Overview of validation process. For each PDB entry, the relevant residues are detected based on their name (3-letter code) and number of atoms. Then, for each validated residue, the corresponding model is retrieved from wwPDB CCD, and each motif of this residue is validated against the model. ValidatorDB is then built as the collection of validation results for all motifs of all residues in all PDB entries. | ||
B) Example of validation for a | B) Example of validation for a sialic acid (SIA) motif in the PDB entry 4jtv. The validated residue SIA is extracted from PDB entry 4jtv in the form of an input motif, which contains all the atoms of the validated residue, together with all atoms found within one or two bonds of any atom from the validated residue (surroundings). By superimposing the input motif and model residue, the validated motif results as the subset of atoms in the input motif which correspond to atoms in the model residue. Evaluation of the validated motif (which atoms are present and where, compared to the model residue) produces the validation results. | ||
C) Typical validation results for a | C) Typical validation results for a SIA motif. If the validated motif contains all atoms at their expected positions, it is marked as correct (green). If any atoms are missing or the motif contains serious issues preventing validation, the validated motif is marked as incomplete (red). If some atoms have wrong chirality, the validated motif is marked accordingly (dark yellow). If any unusual features are detected, the validated motif is marked with a warnings (not shown). The full list of graphical results is available [[ValidatorDB:Analysis_of_Results | here]]. | ||
Latest revision as of 08:07, 21 October 2014
A) Overview of validation process. For each PDB entry, the relevant residues are detected based on their name (3-letter code) and number of atoms. Then, for each validated residue, the corresponding model is retrieved from wwPDB CCD, and each motif of this residue is validated against the model. ValidatorDB is then built as the collection of validation results for all motifs of all residues in all PDB entries.
B) Example of validation for a sialic acid (SIA) motif in the PDB entry 4jtv. The validated residue SIA is extracted from PDB entry 4jtv in the form of an input motif, which contains all the atoms of the validated residue, together with all atoms found within one or two bonds of any atom from the validated residue (surroundings). By superimposing the input motif and model residue, the validated motif results as the subset of atoms in the input motif which correspond to atoms in the model residue. Evaluation of the validated motif (which atoms are present and where, compared to the model residue) produces the validation results.
C) Typical validation results for a SIA motif. If the validated motif contains all atoms at their expected positions, it is marked as correct (green). If any atoms are missing or the motif contains serious issues preventing validation, the validated motif is marked as incomplete (red). If some atoms have wrong chirality, the validated motif is marked accordingly (dark yellow). If any unusual features are detected, the validated motif is marked with a warnings (not shown). The full list of graphical results is available here.
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 08:07, 21 October 2014 |  | 3,350 × 3,550 (2.41 MB) | Lukas (talk | contribs) | |
| 12:54, 26 August 2014 | 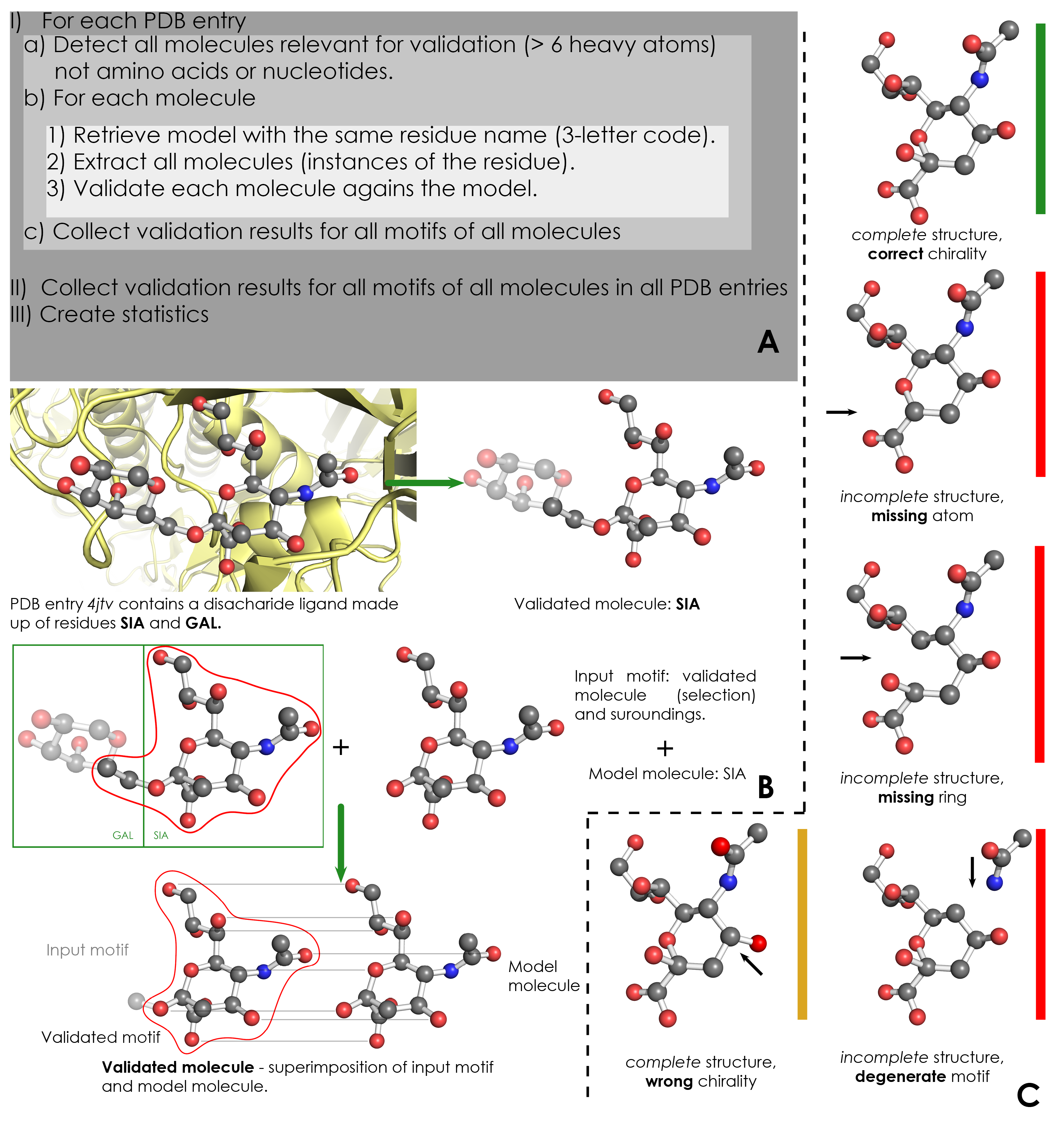 | 3,350 × 3,550 (2.41 MB) | Lukas (talk | contribs) | ||
| 12:17, 26 August 2014 | 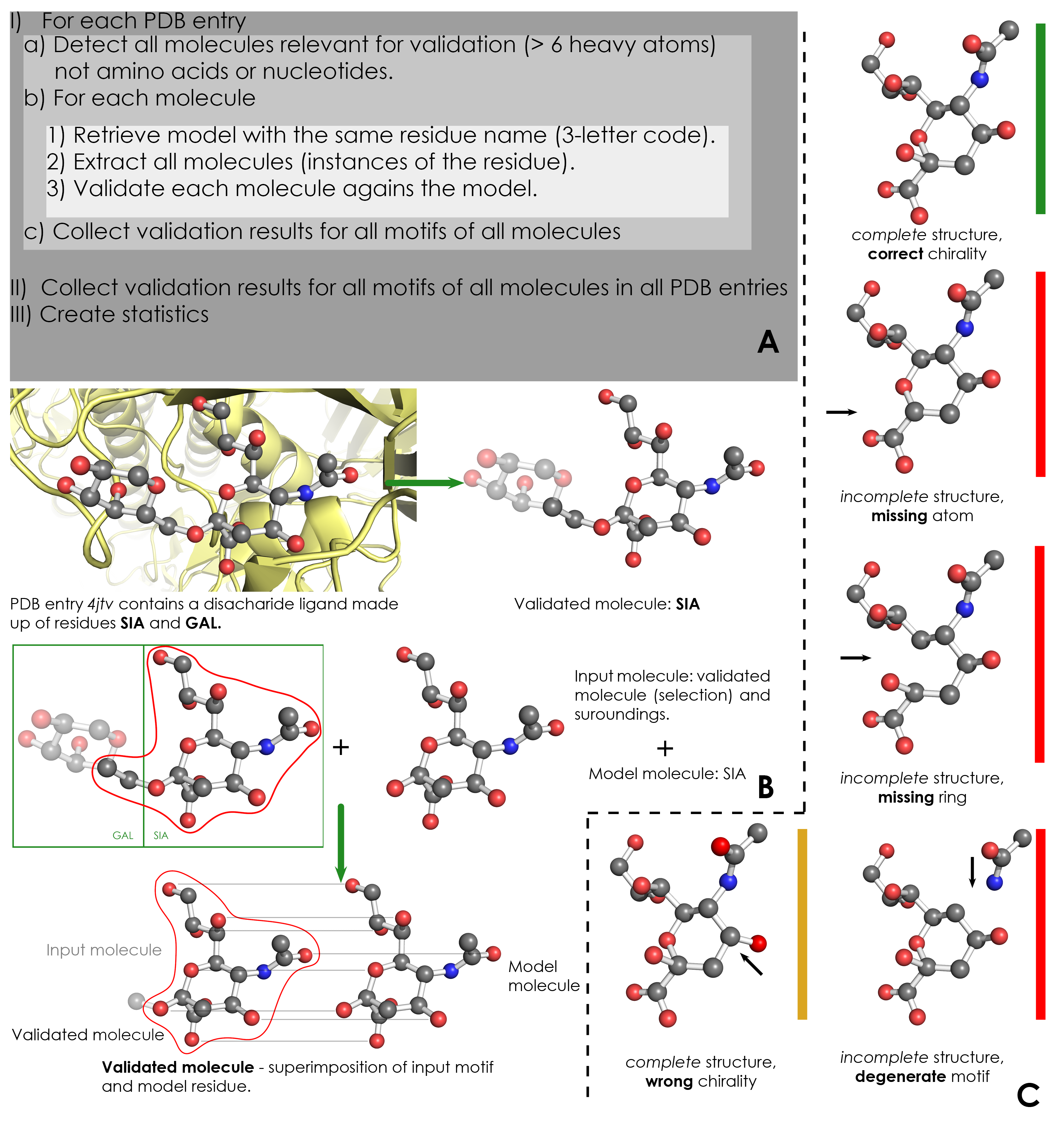 | 3,350 × 3,550 (2.41 MB) | Lukas (talk | contribs) | ||
| 12:16, 26 August 2014 | 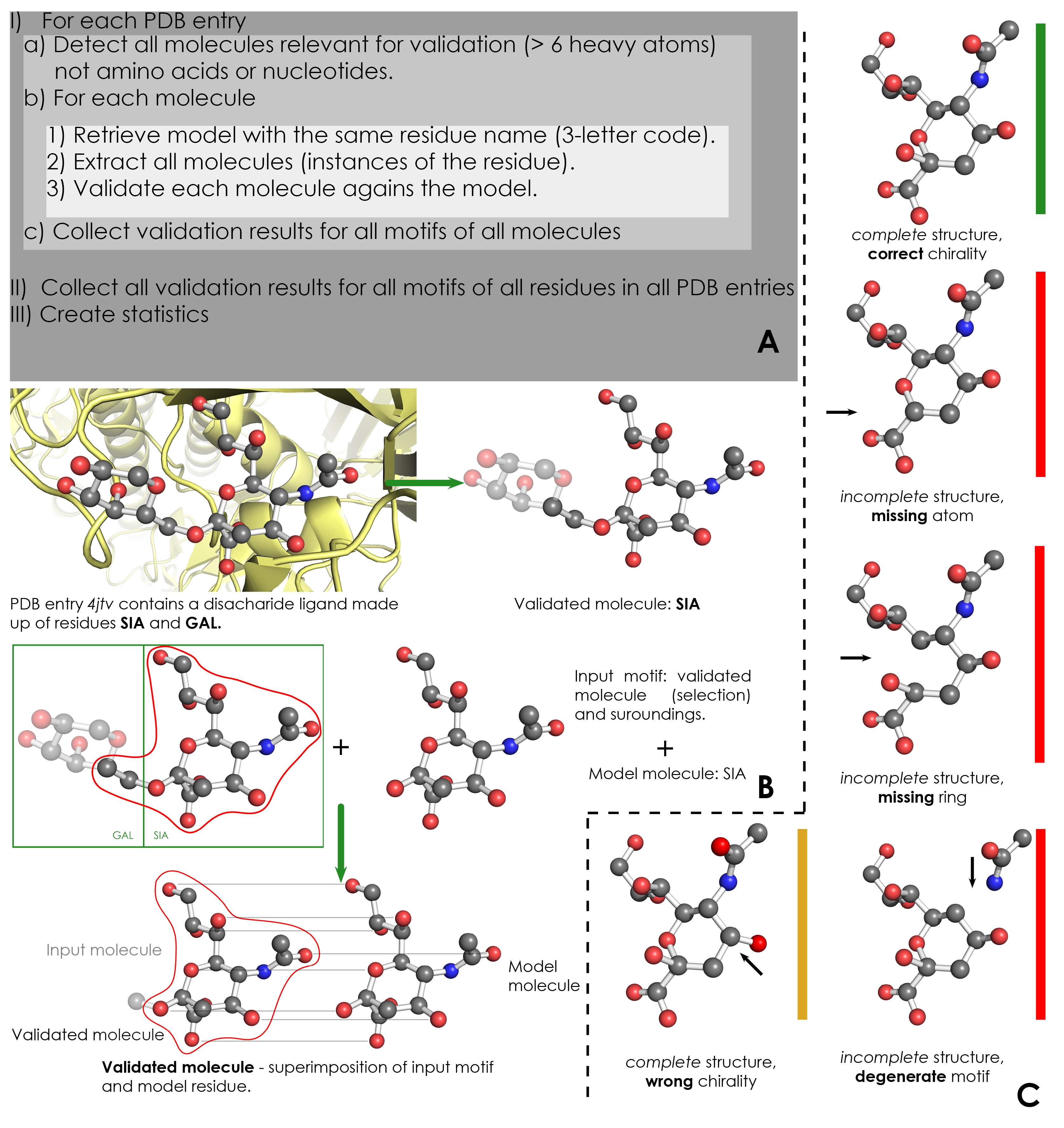 | 3,350 × 3,550 (2.4 MB) | Lukas (talk | contribs) | ||
| 09:51, 14 August 2014 | 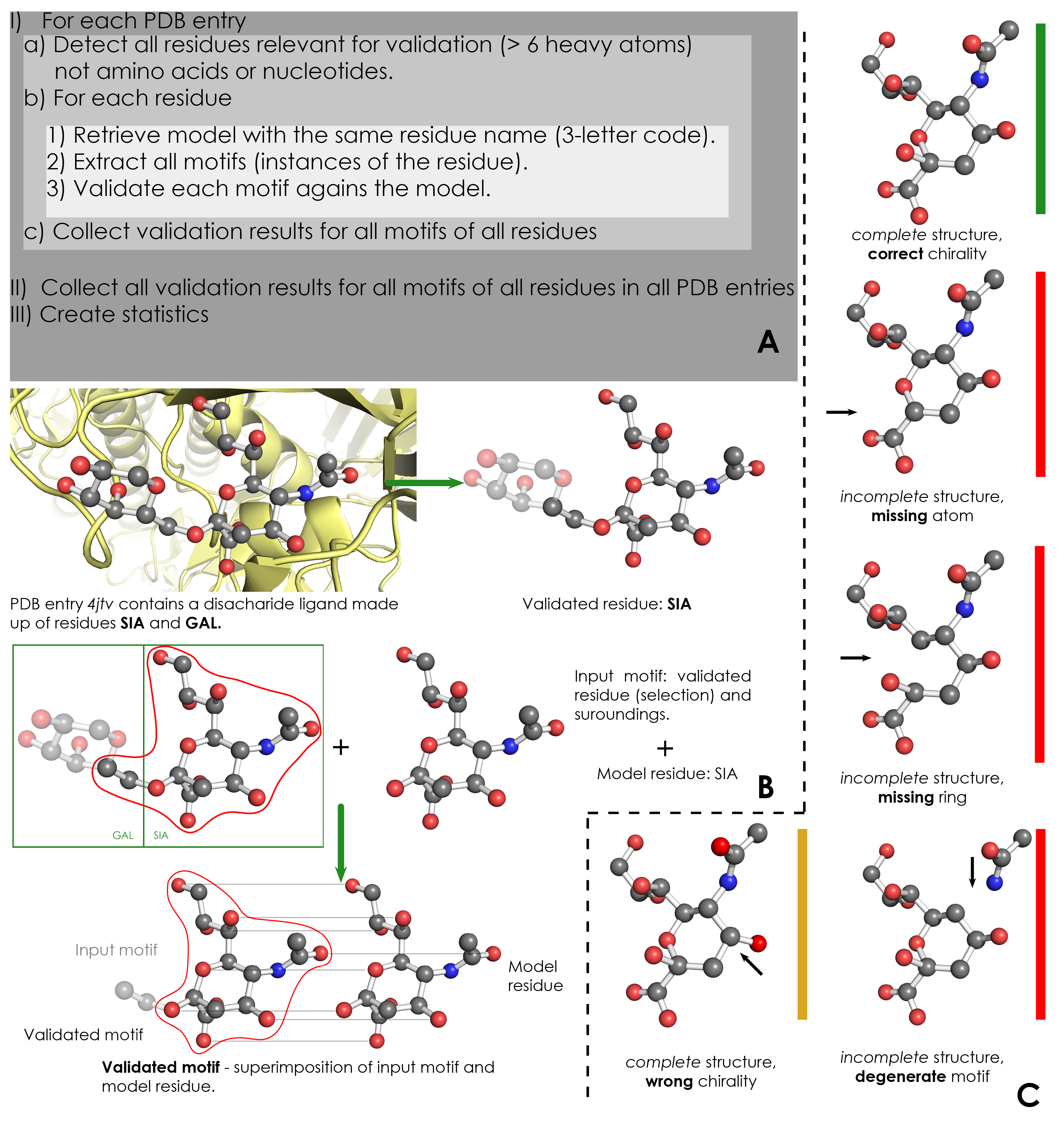 | 3,350 × 3,550 (2.4 MB) | Lukas (talk | contribs) | ||
| 15:13, 12 August 2014 | 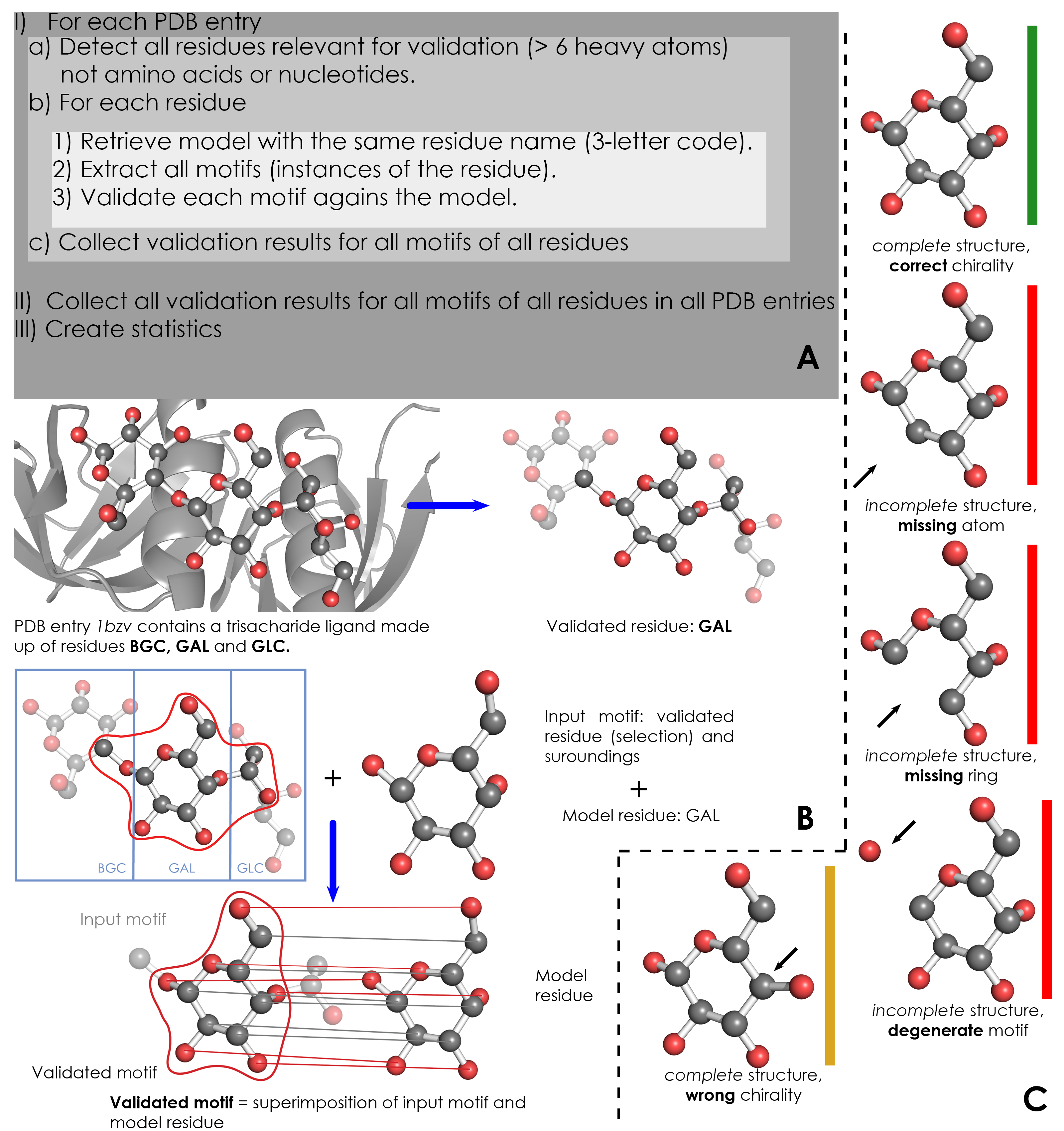 | 3,200 × 3,400 (2.18 MB) | Lukas (talk | contribs) | ||
| 15:15, 5 August 2014 | 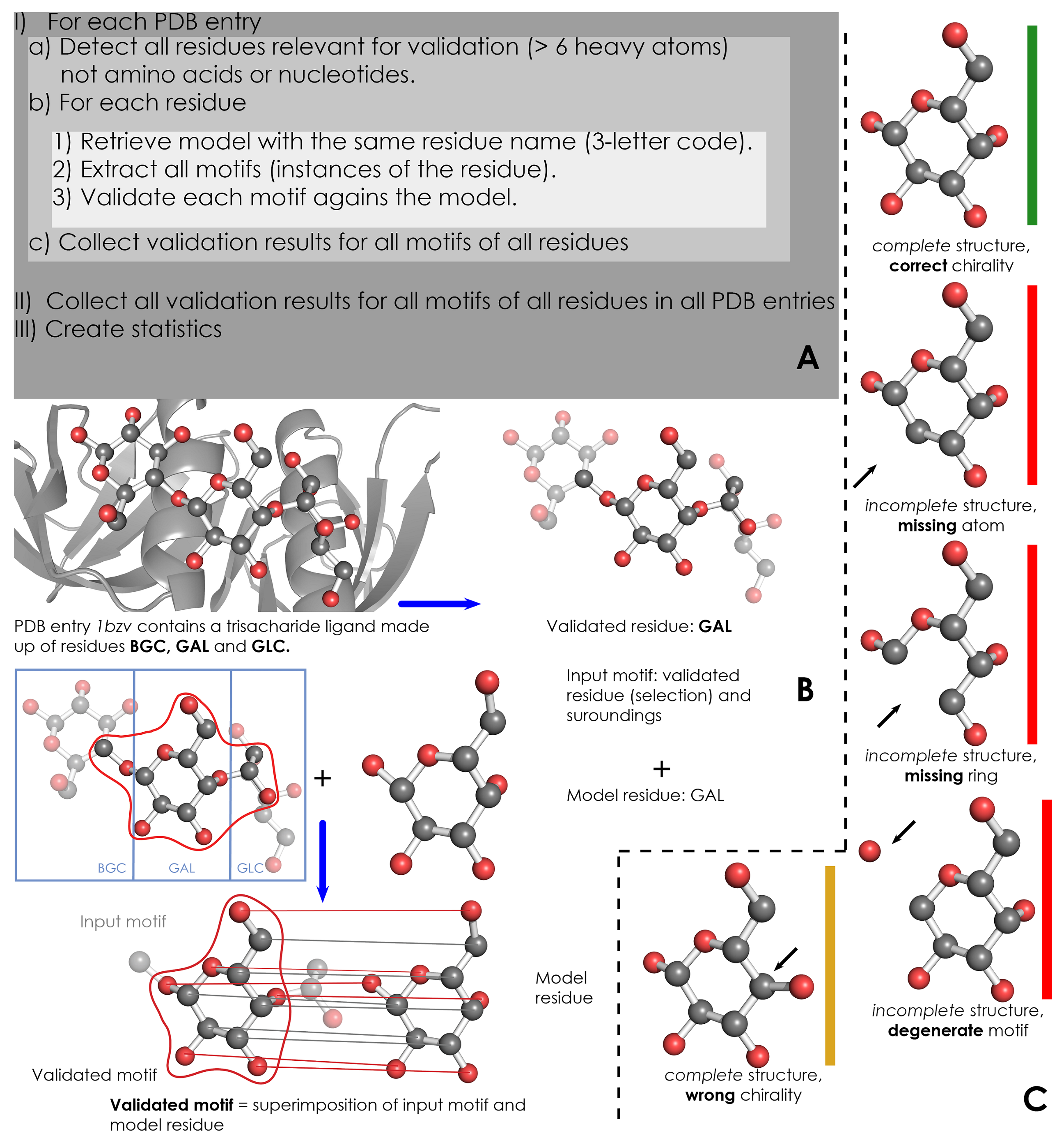 | 2,592 × 2,754 (1.9 MB) | Lukas (talk | contribs) | ||
| 16:04, 10 July 2014 | 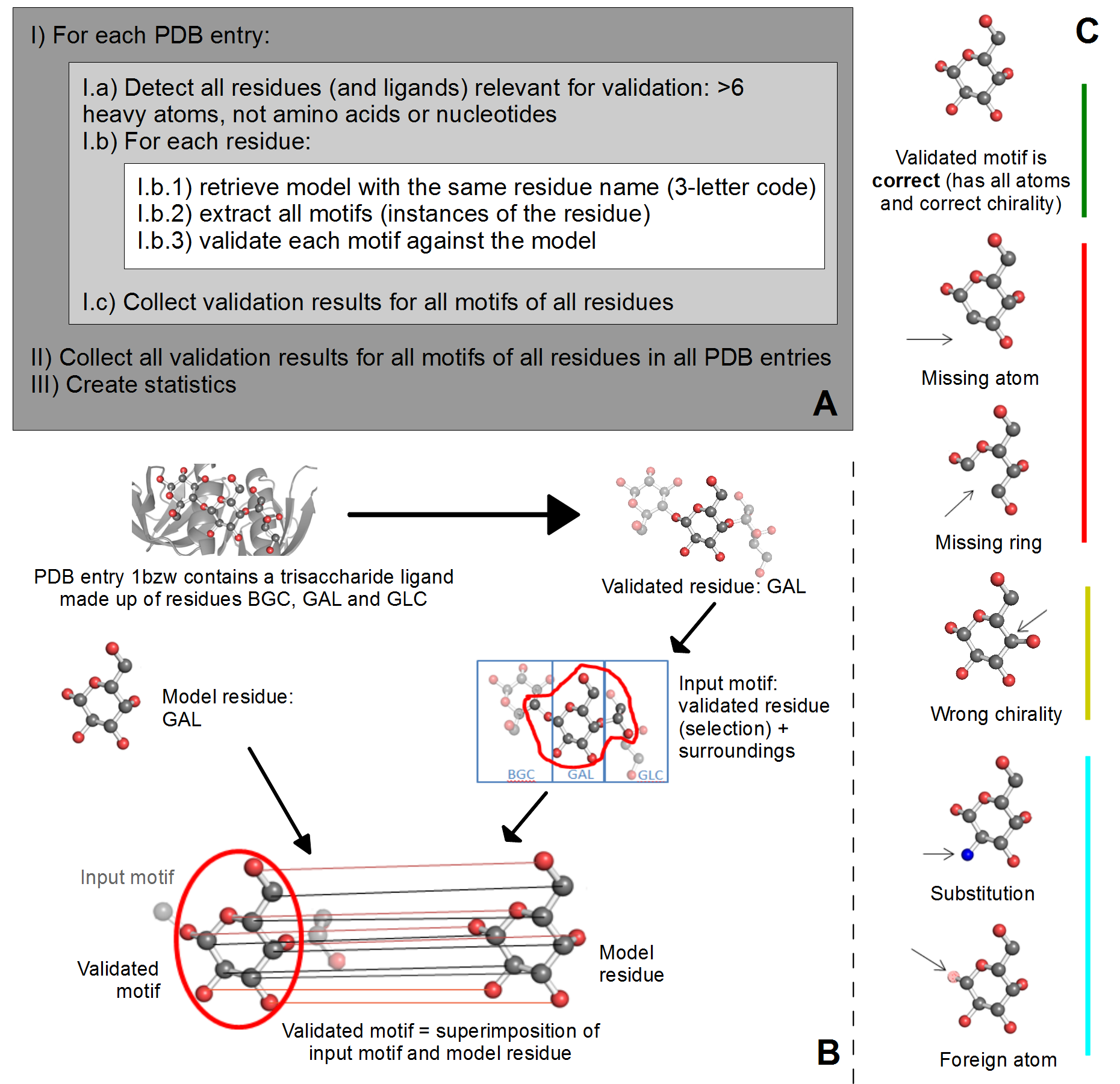 | 1,860 × 1,800 (634 KB) | Deepti (talk | contribs) | ||
| 15:10, 10 July 2014 | 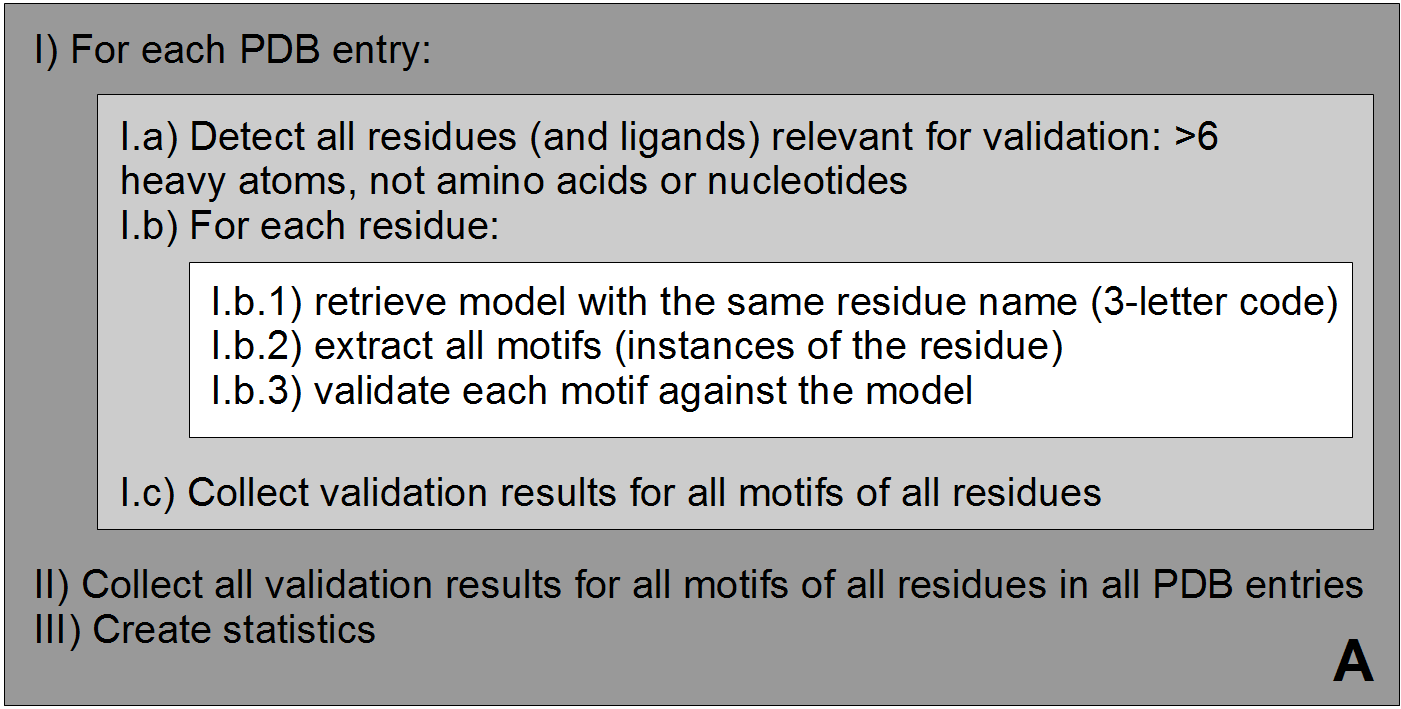 | 1,401 × 709 (51 KB) | Deepti (talk | contribs) | ||
| 13:49, 23 June 2014 | 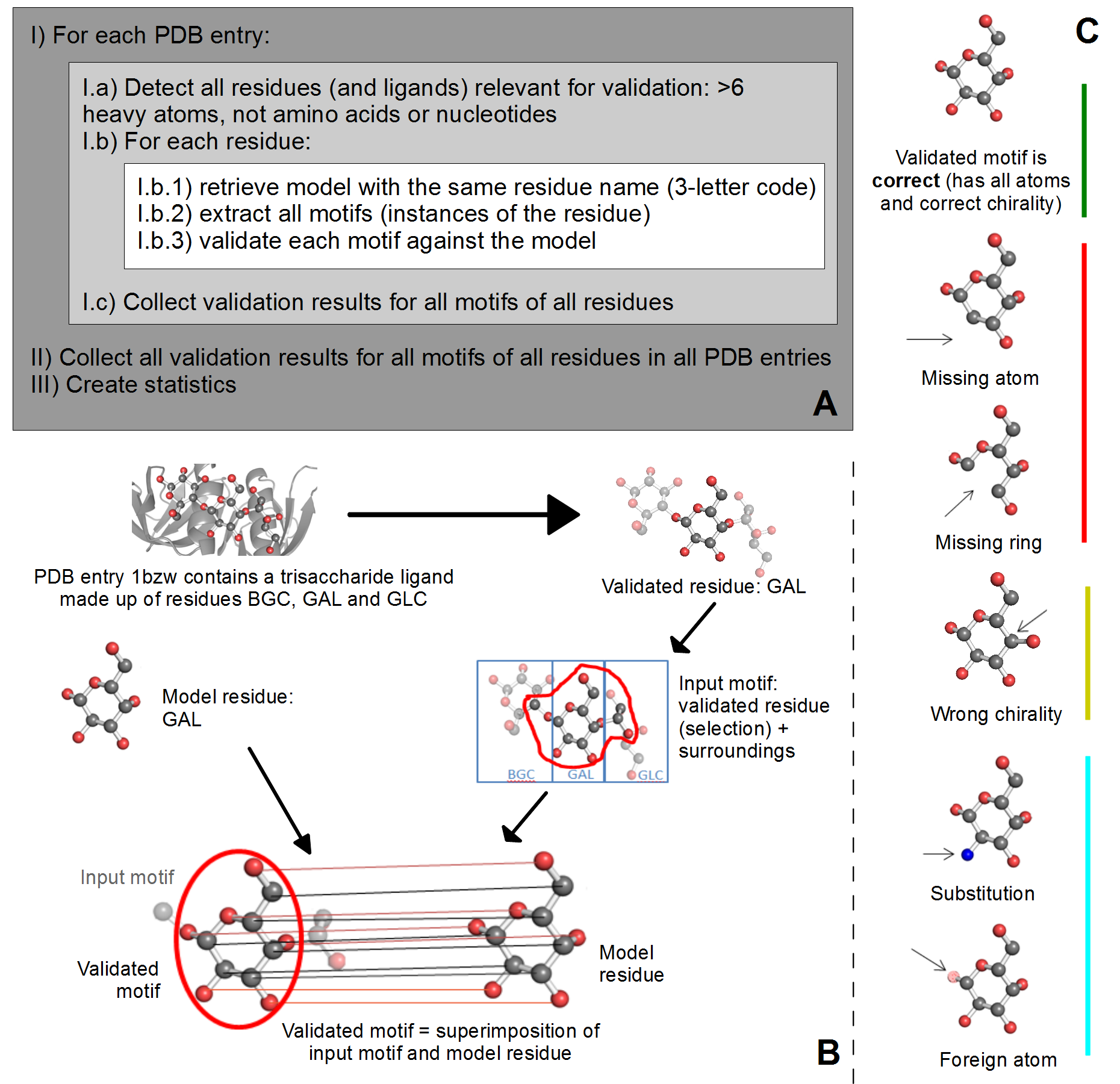 | 1,860 × 1,800 (634 KB) | Deepti (talk | contribs) |
You cannot overwrite this file.
File usage
The following page uses this file: