NEEMP:Reports: Difference between revisions
No edit summary |
No edit summary |
||
| (46 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
Along with '''NEEMP''' we provide an handy python script to generate charge correlation graphs and quality assay reports, named '''''nut-report.py'''''. | |||
< | |||
'''''<u>SCRIPT BASIC USAGE:</u>''''' | |||
''Step 1:'' | |||
* Generation of ''chg-stats-out-file'' in which for each atom the difference between the ''reference QM charge'' and the ''EEM charge'' is calculated ('''''figure 5'''''). Such a file can be obtained running '''NEEMP''' in ''quality validation'' mode with the option <code>--chg-stats-out-file</code> (see [[NEEMP:Examples#Example 8 - Quality validation | example]] or [[NEEMP:Modes#Quality validation | quality validation]] for details). | |||
''Step 2:'' | |||
* Call ''nut-report.py'' with the above-mentioned file | |||
<code>./nut-report.py chg-stats-out-file </code> | |||
'''''Requirements:''''' | |||
* Python ([https://www.python.org/ https://www.python.org/]) | |||
[ | * R ([https://www.r-project.org/ https://www.r-project.org/]) | ||
'''NB''': the script has been tested for both Python 2.7 and Python 3.4, as well as for R 3.2. | |||
<br style="clear:both" /> | |||
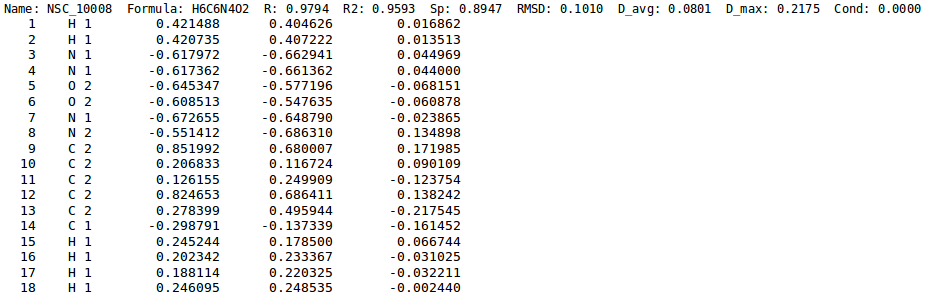
[[File:chg_stats.png | thumb | 900px | center | '''''Figure 5:''''' Close-up from charge statistics file. Along with statistics for each molecule, ''ab-initio'' charges (3rd column), ''EEM'' charges (4th column) and their difference (5th column) are also printed out.]]. | |||
'' | Once the script has been executed, a bunch of output files are generated in the ''chg-stats-out-file'' directory. In particular: | ||
* '' | * '''''csv''''' files containing per-atom charge information and values for several performance evaluating metrics for each atomic type and molecule | ||
* '' | * '''''png''''' files displaying the charge correlation graphs for the whole set and for each atomic type | ||
* '' | * '''''html''''' file gathering together all the previous information in an interactive and more easily readable report page | ||
'''''Figure 6''''' and '''''Figure 7''''' compare few emblematic results extracted from the quality report files for two distinct '''NEEMP''' runs: the first evaluating a parameter set generated using the '''LR''' approach, meanwhile the second a parameter set generated using the '''DE-MIN''' approach. In both cases the training set is ''set02.sdf''. | |||
The following links provide access to the full reports: | |||
* | * [http://www.fi.muni.cz/~xracek/neemp/reports/set02_de/stats_set02_de-report.html set02_DE-MIN] | ||
* [http://www.fi.muni.cz/~xracek/neemp/reports/set02_lr/stats_set02_lr-report.html set02_LR] | |||
{| class="wikitable" border="1" style="margin: | {| class="wikitable" border="1" style="margin-left: auto; margin-right: auto;" width="650px" | ||
|- | |- | ||
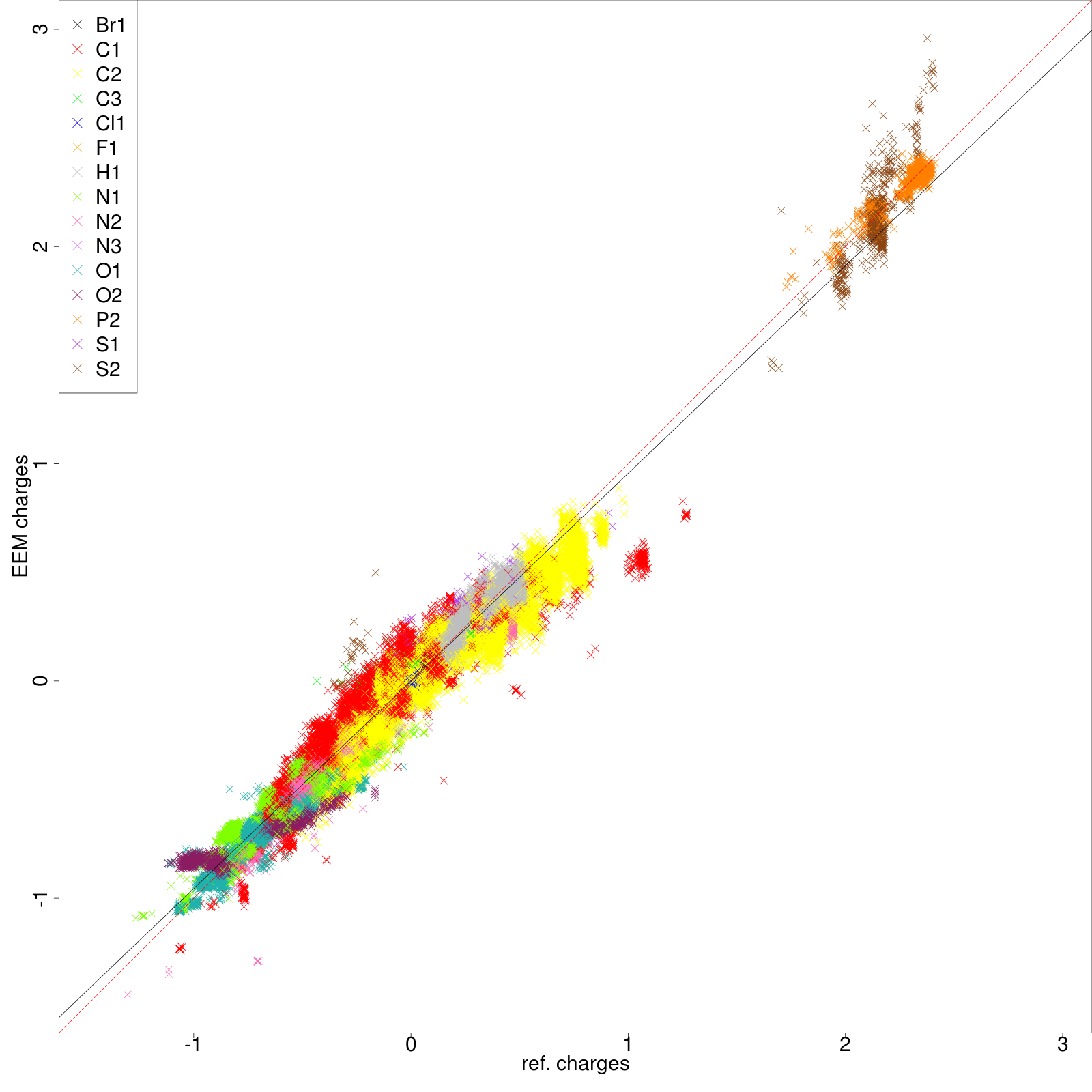
| [[File: | | [[File:stats_set02_de-summary.png| 520px]] | ||
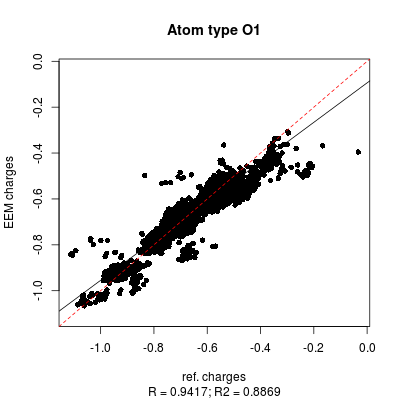
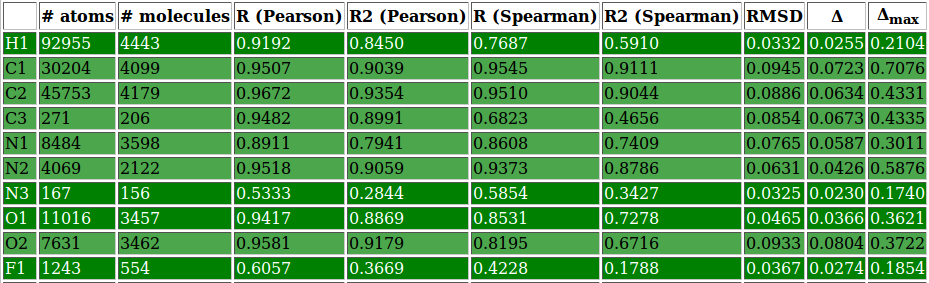
| [[File: | | [[File:stats_set02_de-O1.png| center |400px]] [[File:set02_de_table.png| 650px]] | ||
|- | |- | ||
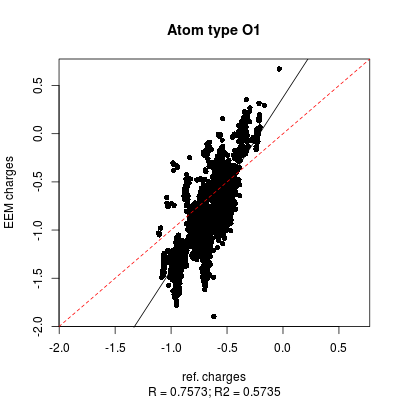
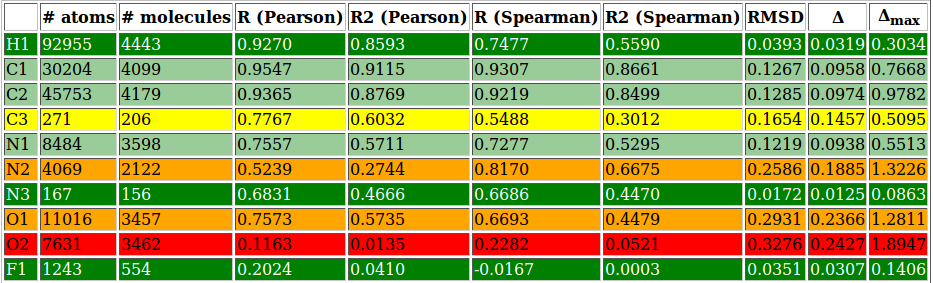
|colspan=2 |'''''Figure 2:''''' | |colspan=2|'''''Figure 6:''''' <u>Left side</u>: charge correlation graph for the whole set. <u>Upper right side</u>: example of charge correlation graph for a single atomic type (in this case oxygen presenting only single bonds). <u>Lower right side</u>: detail from the atomic types summary table, in which each row is coloured according to the ''RMSD'' column (red: high value, green: low value). In this particular case it is evident the high quality performance of the validated parameter set. | ||
|- | |||
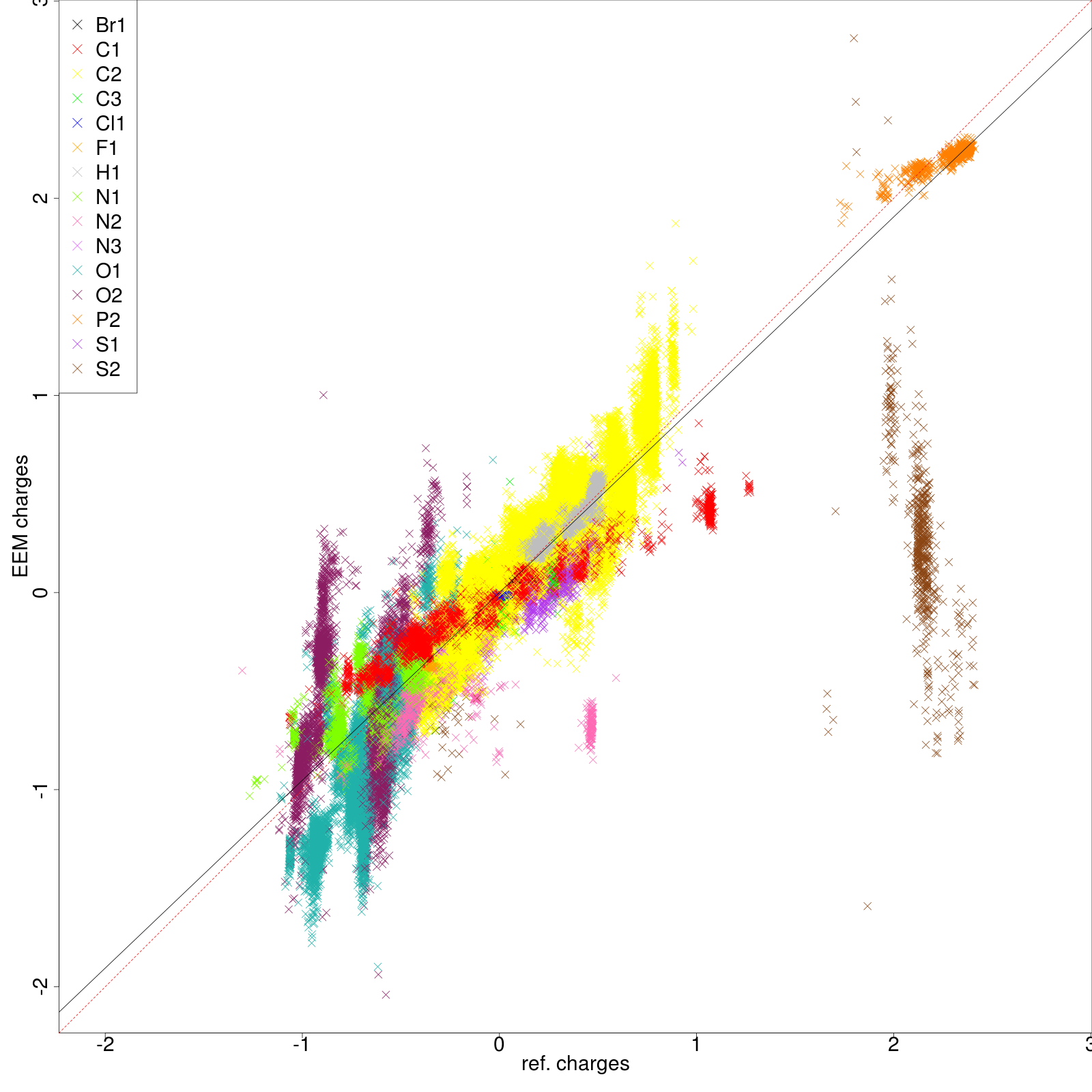
| [[File:stats_set02_lr-summary.png| 520px]] | |||
| [[File:stats_set02_lr-O1.png| center |400px]] [[File:set02_lr_table.png| 650px]] | |||
|- | |||
|colspan=2|'''''Figure 7:''''' For a description of the figure layout refer to '''''figure 6'''''. It can be easily seen how the performance of the submitted parameter set is utterly poor. In specific the correlation graphs help to visualize the low degree of dependency between the ''EEM charges'' and the ''QM charges''. Meanwhile the summary table provides the actual values of several statistical metrics and, as can be seen from the colouring pattern, the ''RMSD'' values are generally higher than the previous case. | |||
|} | |} | ||
Latest revision as of 07:42, 1 July 2016
Along with NEEMP we provide an handy python script to generate charge correlation graphs and quality assay reports, named nut-report.py.
SCRIPT BASIC USAGE:
Step 1:
- Generation of chg-stats-out-file in which for each atom the difference between the reference QM charge and the EEM charge is calculated (figure 5). Such a file can be obtained running NEEMP in quality validation mode with the option
--chg-stats-out-file(see example or quality validation for details).
Step 2:
- Call nut-report.py with the above-mentioned file
./nut-report.py chg-stats-out-file
Requirements:
- Python (https://www.python.org/)
NB: the script has been tested for both Python 2.7 and Python 3.4, as well as for R 3.2.
.
Once the script has been executed, a bunch of output files are generated in the chg-stats-out-file directory. In particular:
- csv files containing per-atom charge information and values for several performance evaluating metrics for each atomic type and molecule
- png files displaying the charge correlation graphs for the whole set and for each atomic type
- html file gathering together all the previous information in an interactive and more easily readable report page
Figure 6 and Figure 7 compare few emblematic results extracted from the quality report files for two distinct NEEMP runs: the first evaluating a parameter set generated using the LR approach, meanwhile the second a parameter set generated using the DE-MIN approach. In both cases the training set is set02.sdf.
The following links provide access to the full reports: