PatternQuery:Atlas: Difference between revisions
Initial fill |
|||
| (2 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{TOC limit|2}} | |||
=DNA= | |||
=RNA kink-turn motif (Kt-7)= | =RNA= | ||
==RNA kink-turn motif (Kt-7)== | |||
[[Image:PatternQuery-Atlas-Kt-7.png|200px|thumb|left|Example from ''1ffk'' protein.]] | [[Image:PatternQuery-Atlas-Kt-7.png|200px|thumb|left|Example from ''1ffk'' protein.]] | ||
| Line 20: | Line 21: | ||
=Protein= | |||
= | ==C<sub>2</sub>H<sub>2</sub> Zinc finger== | ||
[[Image:PatternQuery-Atlas-C2H2.png|200px|thumb|left|Example from ''4r2a'' protein.]] | [[Image:PatternQuery-Atlas-C2H2.png|200px|thumb|left|Example from ''4r2a'' protein.]] | ||
| Line 39: | Line 40: | ||
<br style="clear:both" /> | <br style="clear:both" /> | ||
< | The classical C<sub>2</sub>H<sub>2</sub> zinc finger domain is composed of a simple ββα fold, which is stabilized by a zinc ion coordinated by two histidine and two cysteine residues. The fold is often described by the pattern of X<sub>2</sub>-C-X<sub>2-4</sub>-C-X<sub>12</sub>-H-X<sub>3-5</sub>-H, where X stands for any amino acid, C is cysteine and H is histidine. | ||
</ | |||
==Rubredoxin binding site== | |||
[[Image:PatternQuery-Rubredoxin.png|200px|thumb|right|Example from ''1b13'' protein.]] | |||
===PQ expression=== | |||
<syntaxhighlight lang="python"> | |||
Atoms("Fe").AmbientResidues(2.5).Filter(lambda l: l.Count(Residues("Cys")) == 4) | |||
</syntaxhighlight> | |||
<br style="clear:both" /> | |||
Rubredoxin active site contains an iron ion which is coordinated by the sulfurs of four conserved cysteine residues forming an almost regular tetrahedron. PQ expression identifies Fe atom coordinated by 4 cysteins. | |||
=PA-IIL lectin | ==PA-IIL lectin binding site== | ||
[[Image:PatternQuery-Intro1.png|200px|thumb|left|Example from ''1gzt'' protein.]] | [[Image:PatternQuery-Intro1.png|200px|thumb|left|Example from ''1gzt'' protein.]] | ||
| Line 59: | Line 72: | ||
<br style="clear:both" /> | <br style="clear:both" /> | ||
The PQ expression searches for 2 calcium ions at most 4Å apart, and all the residues with direct interaction with either of these ions. Furthermore, just the molecular patterns containing a residue with a furan or pyran ring were preserved. Additionally, nucleotides are filtered out as well. | |||
Latest revision as of 15:26, 5 August 2015
DNA
[edit]RNA
[edit]RNA kink-turn motif (Kt-7)
[edit]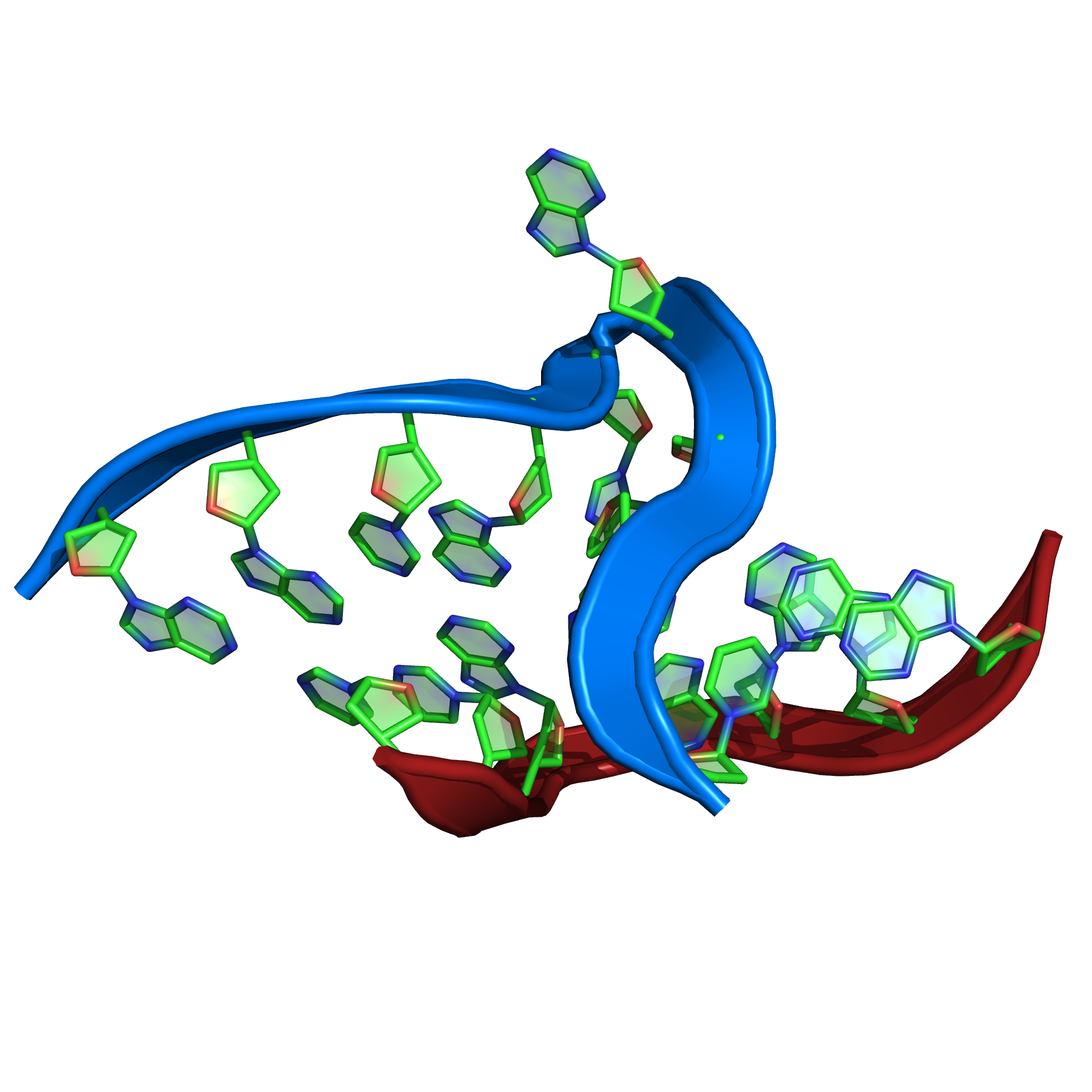
PQ expression
[edit] Near(4,
RegularMotifs(’GGCGAAGAAC’, Type=’Nucleotide’),
RegularMotifs(’GGGAGCC’, Type=’Nucleotide’))
Some description goes here
Protein
[edit]C2H2 Zinc finger
[edit]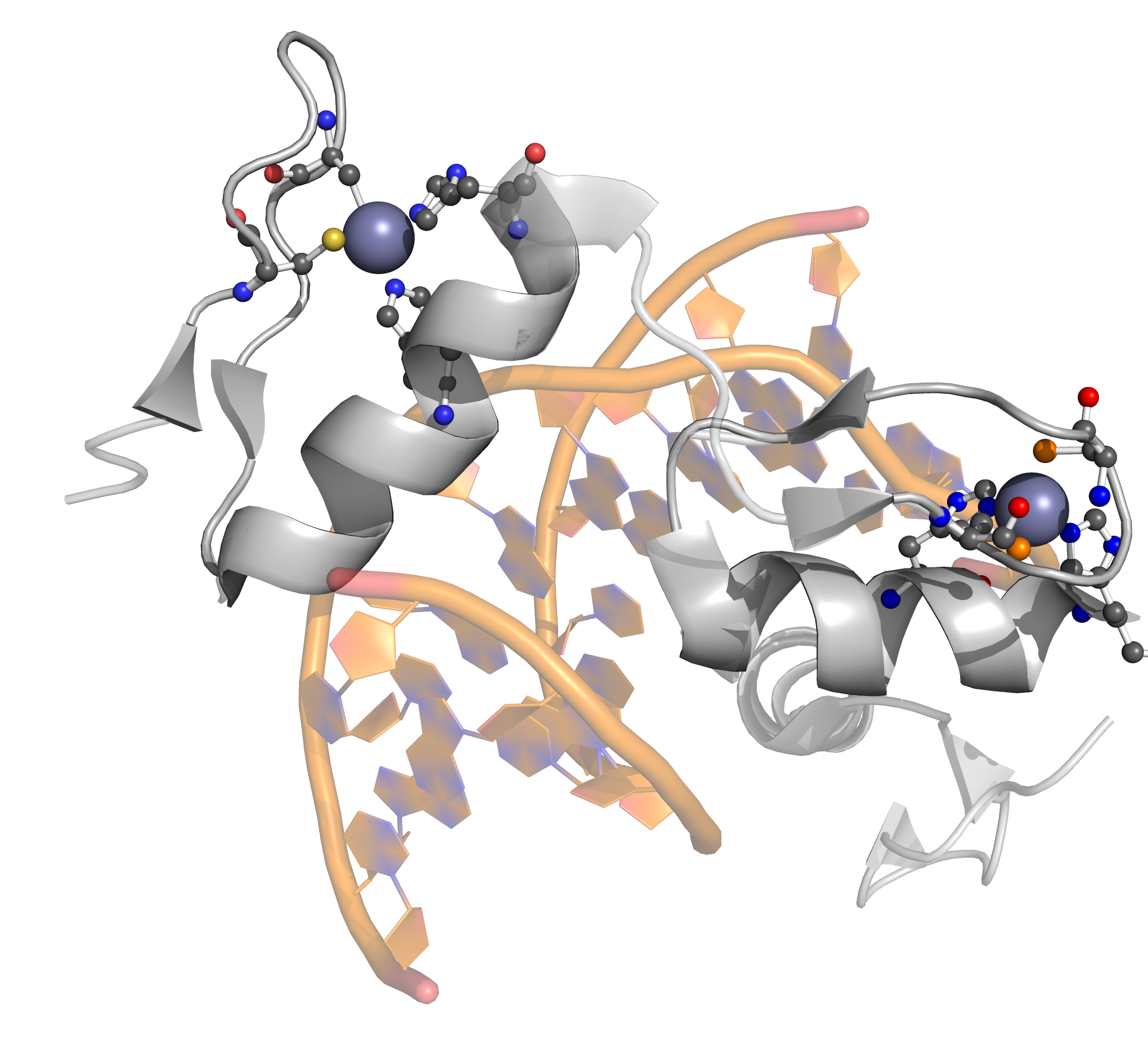
PQ expression
[edit]RegularMotifs('.{2}C.{2,4}C.{12}H.{3,5}H').
ConnectedAtoms(1).
Filter(lambda m:
m.Find(Atoms('Zn').
ConnectedResidues(1).
Filter(lambda n:
(n.Count(Residues('Cys')) == 2) & (n.Count(Residues('His')) == 2))).
SeqCount() > 0)
The classical C2H2 zinc finger domain is composed of a simple ββα fold, which is stabilized by a zinc ion coordinated by two histidine and two cysteine residues. The fold is often described by the pattern of X2-C-X2-4-C-X12-H-X3-5-H, where X stands for any amino acid, C is cysteine and H is histidine.
Rubredoxin binding site
[edit]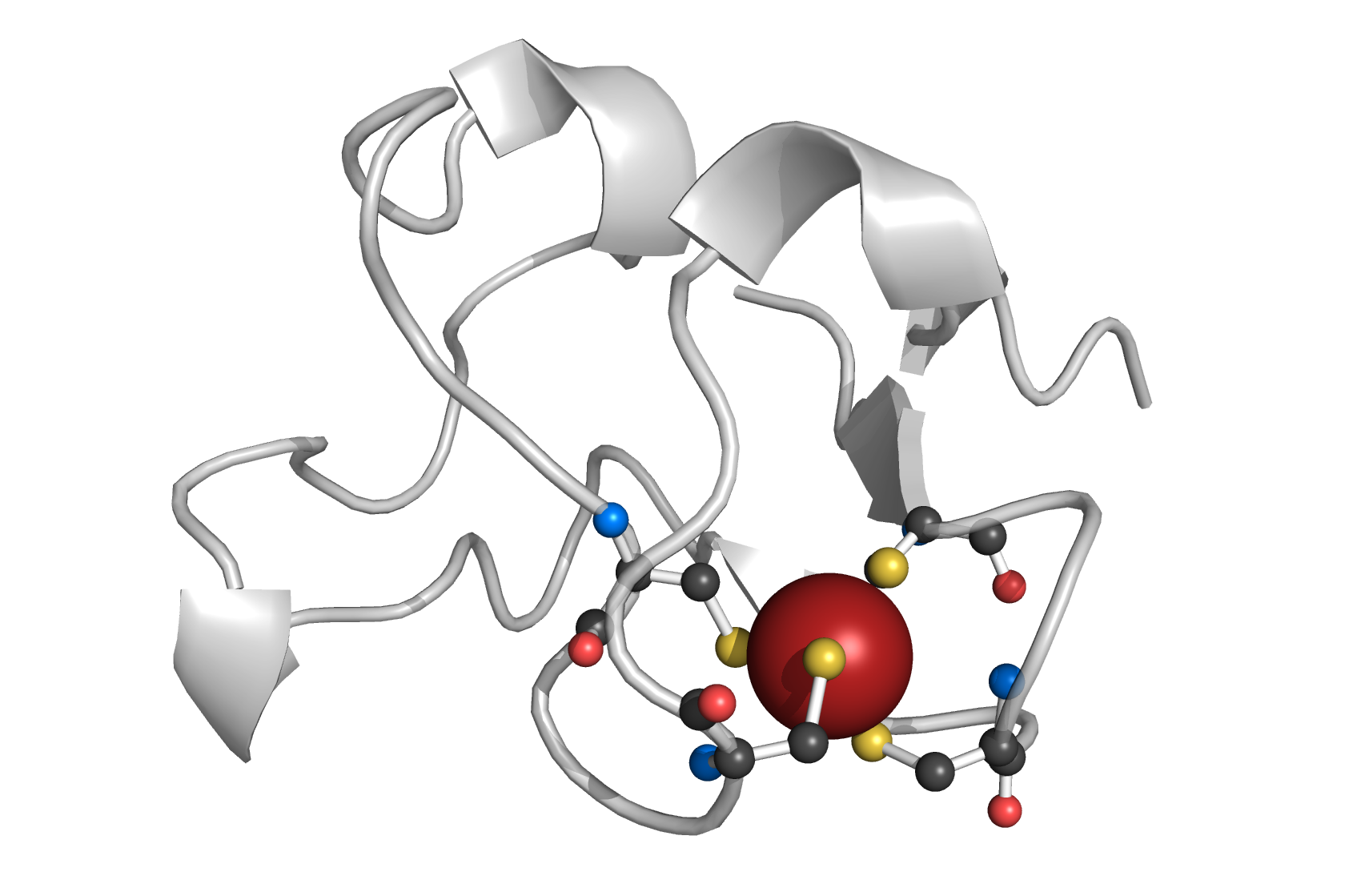
PQ expression
[edit]Atoms("Fe").AmbientResidues(2.5).Filter(lambda l: l.Count(Residues("Cys")) == 4)
Rubredoxin active site contains an iron ion which is coordinated by the sulfurs of four conserved cysteine residues forming an almost regular tetrahedron. PQ expression identifies Fe atom coordinated by 4 cysteins.
PA-IIL lectin binding site
[edit]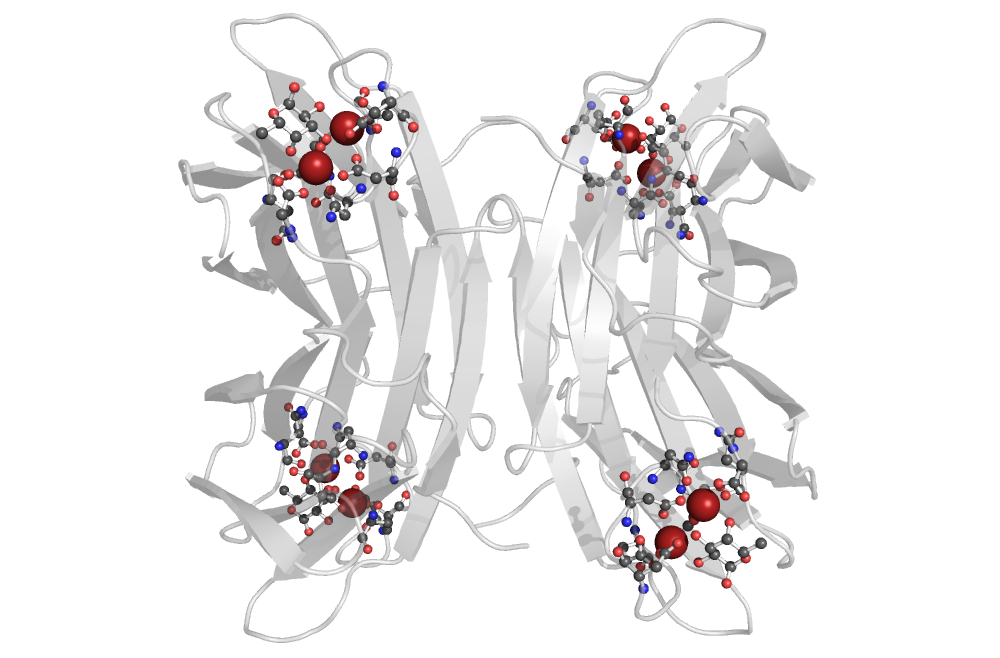
PQ expression
[edit]
Near(4, Atoms("Ca"), Atoms("Ca"))
.ConnectedResidues(1)
.Filter(lambda l:
l.Count(Or(Rings(5 * ["C"] + ["O"]), Rings(4 * ["C"] + ["O"]))) > 0)
.Filter(lambda l: l.Count(Atoms("P")) == 0)
The PQ expression searches for 2 calcium ions at most 4Å apart, and all the residues with direct interaction with either of these ions. Furthermore, just the molecular patterns containing a residue with a furan or pyran ring were preserved. Additionally, nucleotides are filtered out as well.