LiteMol:Functionality: Difference between revisions
Appearance
→Classic features: mjc |
|||
| (4 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
=Classic features= | =Classic features= | ||
* Supports all major structural data formats ([http://mmcif.wwpdb.org/ PDBx/mmcif], [https://www.wwpdb.org/documentation/file-format PDB], [http://pubs.acs.org/doi/pdf/10.1021/ci00007a012 SDF]). | * Supports all major structural data formats ([http://mmcif.wwpdb.org/ PDBx/mmcif], [https://www.wwpdb.org/documentation/file-format PDB], [http://pubs.acs.org/doi/pdf/10.1021/ci00007a012 SDF]). | ||
* All common visualization models ('balls- | * All common visualization models ('balls-and-sticks'/cartoon/Cα-trace/VDW spheres/surfaces). | ||
* Displays | * Displays electron densities for X-ray crystallographic structures and electronic potential maps for cryo-EM models. | ||
* Available as a standalone viewer or as a library for embedding into third party applications. | * Available as a [https://webchemdev.ncbr.muni.cz/LiteMol/Viewer/ standalone viewer] or as a [https://github.com/dsehnal/LiteMol library} for embedding into third party applications. | ||
[[Image:LiteMol-Models.png|center|800px]] | [[Image:LiteMol-Models.png|center|800px]] | ||
=Advanced | =Advanced features= | ||
* Introduces the [https://github.com/dsehnal/BinaryCIF BinaryCIF] a compression format compatible with standard PDBx/mmCIF format ''greatly reducing'' amount of transferred data. | * Introduces the [https://github.com/dsehnal/BinaryCIF BinaryCIF] a compression format compatible with standard PDBx/mmCIF format ''greatly reducing'' the amount of transferred data. | ||
* [https://webchem.ncbr.muni.cz/CoordinateServer CoordinateServer] allows for streaming only parts of the | * [https://webchem.ncbr.muni.cz/CoordinateServer CoordinateServer] allows for streaming only those parts of the structural model relevant for the user required visualization. This further ''decreases'' the amount of ''transferred data'' and facilitates ''near instant responsiveness'' even for huge structures such as ribosomes and viral capsids. All in your web browser. | ||
* [http://webchem.ncbr.muni.cz/DensityServer DensityServer ] | * [http://webchem.ncbr.muni.cz/DensityServer DensityServer ] allows ''instant access'' to full resolution user determined slices of ''electron density'' (e.g. binding sites) and down sampled surface of the entire biomacromolecular structure. | ||
* Displays [https://www.wwpdb.org/validation/validation-reports | * Displays [https://www.wwpdb.org/validation/validation-reports wwPDB validation reports] and [[ValidatorDB:UserManual | ValidatorDB ligand validation]]. | ||
* Displays ''sequence annotations'' from [http://pfam.xfam.org/ Pfam], [https://www.ebi.ac.uk/interpro/ InterPro], [www.cathdb.info CATH | * Displays ''sequence annotations'' from [http://pfam.xfam.org/ Pfam], [https://www.ebi.ac.uk/interpro/ InterPro], [www.cathdb.info CATH], [http://scop.berkeley.edu/ SCOP], [http://www.uniprot.org/ UniProt]. | ||
* | * Visualizer requires only an up-to-date ''web browser'' with no dependencies required. Runs on all ''mobile devices''. | ||
* Minimal ''memory footprint''. | * Minimal ''memory footprint''. | ||
[[Image:LiteMol-Performance.png|center|800px]] | [[Image:LiteMol-Performance.png|center|800px]] | ||
Latest revision as of 12:18, 5 May 2017
{{#if: LiteMol is a powerful and blazing-fast cross-platform tool for handling 3D macromolecular data in the browser. Try it now! | LiteMol is a powerful and blazing-fast cross-platform tool for handling 3D macromolecular data in the browser. Try it now! |large }}
Classic features
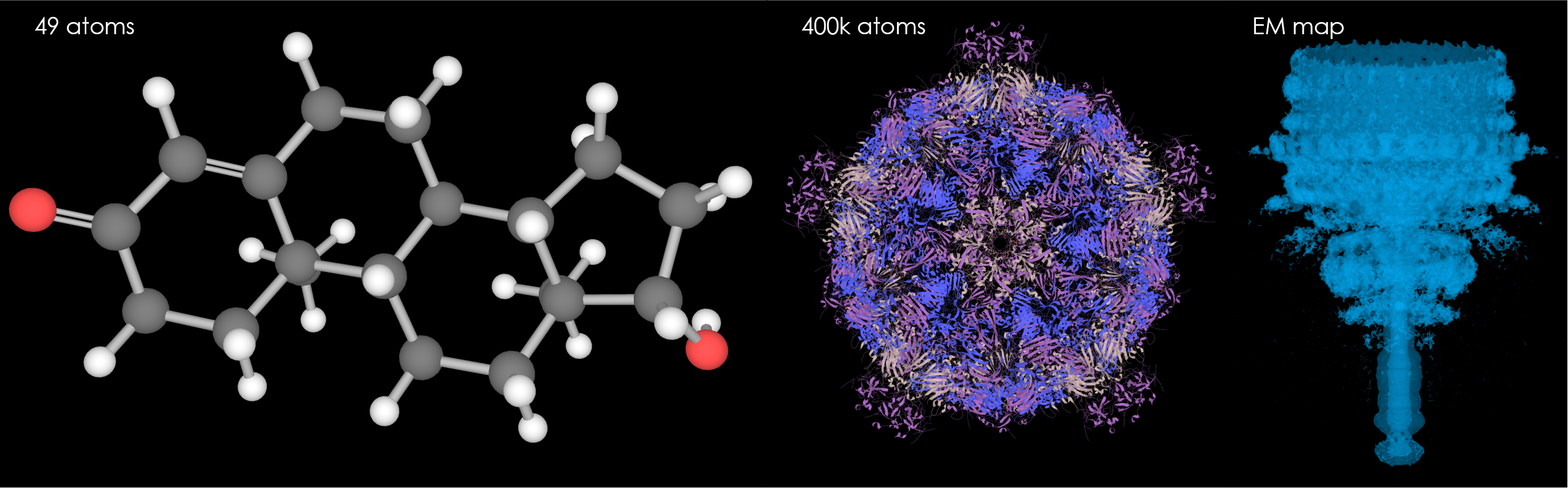
[edit]- Supports all major structural data formats (PDBx/mmcif, PDB, SDF).
- All common visualization models ('balls-and-sticks'/cartoon/Cα-trace/VDW spheres/surfaces).
- Displays electron densities for X-ray crystallographic structures and electronic potential maps for cryo-EM models.
- Available as a standalone viewer or as a [https://github.com/dsehnal/LiteMol library} for embedding into third party applications.
Advanced features
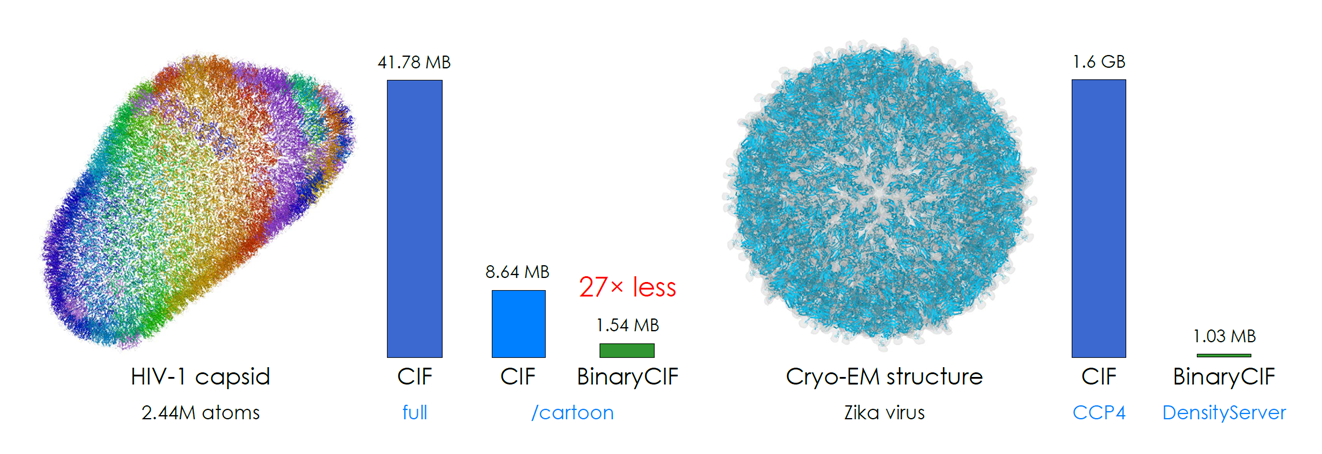
[edit]- Introduces the BinaryCIF a compression format compatible with standard PDBx/mmCIF format greatly reducing the amount of transferred data.
- CoordinateServer allows for streaming only those parts of the structural model relevant for the user required visualization. This further decreases the amount of transferred data and facilitates near instant responsiveness even for huge structures such as ribosomes and viral capsids. All in your web browser.
- DensityServer allows instant access to full resolution user determined slices of electron density (e.g. binding sites) and down sampled surface of the entire biomacromolecular structure.
- Displays wwPDB validation reports and ValidatorDB ligand validation.
- Displays sequence annotations from Pfam, InterPro, [www.cathdb.info CATH], SCOP, UniProt.
- Visualizer requires only an up-to-date web browser with no dependencies required. Runs on all mobile devices.
- Minimal memory footprint.