NEEMP:Modes: Difference between revisions
| (32 intermediate revisions by the same user not shown) | |||
| Line 15: | Line 15: | ||
To compute EEM charges with '''NEEMP''', it is necessary to provide the SDF file with molecules for which the charges will be computed (option <code>--sdf-file</code>) and the PAR file with EEM parameters (option <code>--par-file</code>). EEM charges will be written into the file specified by <code>--chg-out-file</code> option. | To compute EEM charges with '''NEEMP''', it is necessary to provide the SDF file with molecules for which the charges will be computed (option <code>--sdf-file</code>) and the PAR file with EEM parameters (option <code>--par-file</code>). EEM charges will be written into the file specified by <code>--chg-out-file</code> option. | ||
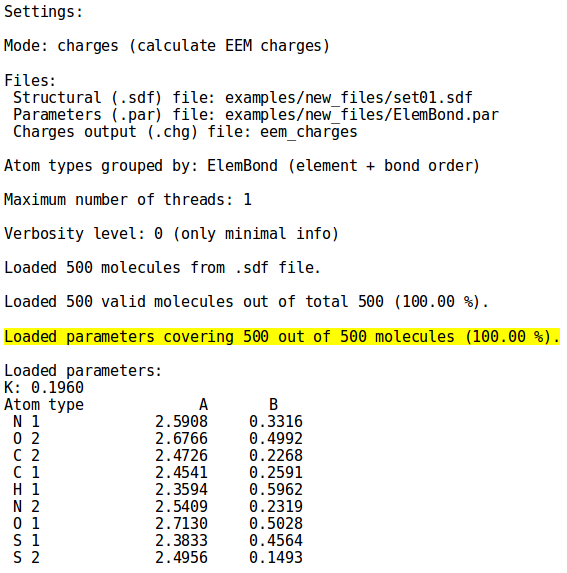
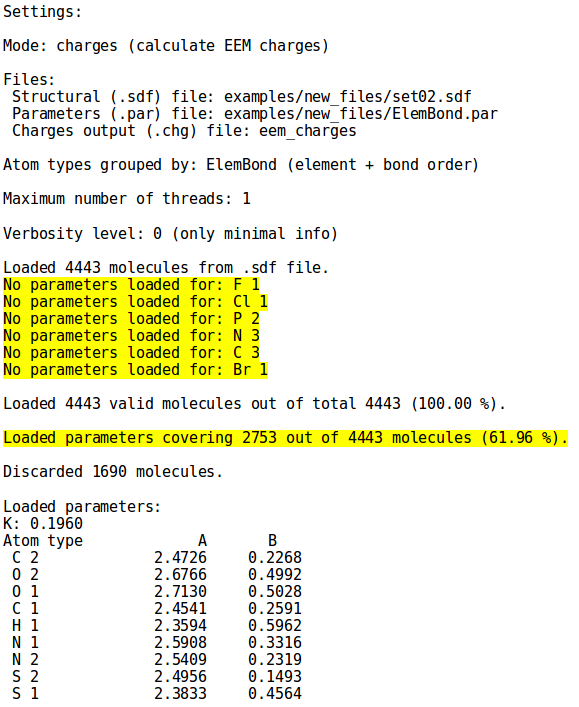
The user should keep in mind that, because of the EEM method itself, the EEM charges cannot be computed for atoms or atomic types for which the parameters are missing. This situation is well depicted by '''''Figure 1''''' | The user should keep in mind that, because of the EEM method itself, the EEM charges cannot be computed for atoms or atomic types for which the parameters are missing. This situation is well depicted by '''''Figure 1'''''. On the left side the parameter set contains values for all the atomic types in ''set01.sdf'', on the other hand in the right side image it is evident how the same parameter set is lacking values for several atomic types present in ''set02.sdf'', leading to the discard of many molecules (for details on the syntax, see the [[NEEMP:Examples#Example 2 - Calculation mode | examples]] or [[NEEMP:Options | option list]] section). | ||
<br style="clear:both" /> | <br style="clear:both" /> | ||
{| class="wikitable" border="1" style="margin: | {| class="wikitable" border="1" style="margin-left: auto; margin-right: auto;" width="650px" | ||
|- | |- | ||
| [[File:chg_new1.png| | | [[File:chg_new1.png| 450px]] | ||
| [[File:chg_new2.png| 450px]] | | [[File:chg_new2.png| 450px]] | ||
|- | |- | ||
|colspan=2 |'''''Figure 1:''''' | |colspan=2 |'''''Figure 1:''''' The lack of parameter values in the parameter set file ''ElemBond.par'' for the atomic types highlighted in the right side image causes the discarding of several molecules from ''set02.sdf'', emphasized by the drop in the covered molecules percentage. | ||
|} | |} | ||
| Line 45: | Line 45: | ||
:<code>-s</code> <code>RMSD</code> or <code>--sort-by</code> <code>RMSD</code> to select the parameter set with the lowest atomic type root mean square difference (avg(RMSDa)) | :<code>-s</code> <code>RMSD</code> or <code>--sort-by</code> <code>RMSD</code> to select the parameter set with the lowest atomic type root mean square difference (avg(RMSDa)) | ||
{| class="wikitable" border="1" style="margin: | {| class="wikitable" border="1" style="margin-left: auto; margin-right: auto;" width="650px" | ||
|- | |- | ||
| [[File:LR_full_R2.png| | | [[File:LR_full_R2.png| 600px]] | ||
| [[File: | | [[File:LR_full_RMSD.png| 600px]] | ||
|- | |- | ||
|colspan=2 |'''''Figure 2:''''' Detailed view of the '''LR''' parametrization settings for two distinct '''NEEMP''' executions differing in the best-performance selection metrics (''R<sup>2</sup>'' in left side image and ''RMSD'' in the right side image). | |colspan=2 |'''''Figure 2:''''' Detailed view of the '''LR''' parametrization settings for two distinct '''NEEMP''' executions differing in the best-performance selection metrics (''R<sup>2</sup>'' in left side image and ''RMSD'' in the right side image). | ||
|} | |} | ||
Additionally, the quality of the parameter set may benefit from the usage of a discarding procedure (<code>-d</code> <code>simple</code> or <code>--discard</code> <code>simple</code>). In fact it has been proved that using a subset of the original training set often leads to EEM charges that better agree with the reference QM charges. For useful examples on the '''LR''' parametrization approach and the discarding procedure see respectively examples [[NEEMP:Examples#Example | Additionally, the quality of the parameter set may benefit from the usage of a discarding procedure (<code>-d</code> <code>simple</code> or <code>--discard</code> <code>simple</code>). In fact it has been proved that using a subset of the original training set often leads to EEM charges that better agree with the reference QM charges. For useful examples on the '''LR''' parametrization approach and the discarding procedure see respectively examples [[NEEMP:Examples#Example 3 - Parametrization mode | 3]]/[[NEEMP:Examples#Example 4 - Parametrization mode k search |4]] and [[NEEMP:Examples#Example 5 - Parametrization mode simple discard|5]]. In particular the latter introduces the <code>--limit-iters</code> and <code>--limit-time</code> options, which modulate the behaviour of the discarding procedure, setting respectively an upper bound in the number of iterations and/or in the execution time. | ||
===Differential Evolution + Minimization (DE-MIN)=== | ===Differential Evolution + Minimization (DE-MIN)=== | ||
| Line 59: | Line 59: | ||
'''''<u>BASIC USAGE:</u>''''' ''./neemp -m params -p de --sdf-file examples/set01.sdf --chg-file examples/charges.chg --par-out-file new_parameter.par'' | '''''<u>BASIC USAGE:</u>''''' ''./neemp -m params -p de --sdf-file examples/set01.sdf --chg-file examples/charges.chg --par-out-file new_parameter.par'' | ||
In parallel we developed an advanced EEM parametrization approach specifically tailored to cope with heterogeneous data, which in the past presented quite a challenge in the EEM parametrization framework. It consists in the combination of ''differential evolution'' method with a local minimization technique (''NEWUOA''), hence the | In parallel we developed an advanced EEM parametrization approach specifically tailored to cope with heterogeneous data, which in the past presented quite a challenge in the EEM parametrization framework. It consists in the combination of ''differential evolution'' method with a local minimization technique (''NEWUOA''), hence the acronym '''DE-MIN'''. To call this approach in '''NEEMP''' use the option <code>--params-method</code> <code>de</code> or <code>-p</code> <code>de</code>. | ||
<br style="clear:both" /> | <br style="clear:both" /> | ||
[[File:gm1.png | thumb | 700px | center | '''''Figure 3:''''' Abridged '''DE-MIN''' approach output for | [[File:gm1.png | thumb | 700px | center | '''''Figure 3:''''' Abridged '''DE-MIN''' approach output for structural file ''examples/set01.sdf'' and charge file ''examples/set01.chg''. In the upper block are displayed the specifics for the '''de-min''' parametrization procedure. Note as the bottom block presents the same structure as for other parametrization approaches and options (compare [[NEEMP:Examples#Example 5 - Parametrization mode simple discard | here]]).]] | ||
The behaviour of the '''DE-MIN''' approach can be modulated by several options, but to make life easier we provide the user with reasonable default values that have been tuned to output optimal parameter set (for a complete view of the options refer to the [[NEEMP:Options | option list]] and the [[NEEMP:Examples#Example | The behaviour of the '''DE-MIN''' approach can be modulated by several options, but to make life easier we provide the user with reasonable default values that have been tuned to output optimal parameter set (for a complete view of the options refer to the [[NEEMP:Options | option list]] and the [[NEEMP:Examples#Example 6 - Parametrization mode DE-MIN | examples]] section). | ||
'''NB''': the default quality evaluating metrics for '''DE-MIN''' approach is ''(avg(RMSDa))''. | '''NB''': the default quality evaluating metrics for '''DE-MIN''' approach is ''(avg(RMSDa))''. | ||
| Line 74: | Line 74: | ||
===Coverage validation=== | ===Coverage validation=== | ||
'''''<u>BASIC USAGE:</u>''''' ''./neemp -m cover -sdf-file examples/ | '''''<u>BASIC USAGE:</u>''''' ''./neemp -m cover -sdf-file examples/set01.sdf --par-file examples/Element.par'' | ||
[[File:cover_mode.png | thumb | 500px | right | '''''Figure 4:''''' Percentage of molecules in set ''examples/ | [[File:cover_mode.png | thumb | 500px | right | '''''Figure 4:''''' Percentage of molecules in set ''examples/set01.sdf'' covered by the displayed parameters set ''examples/Element.par'']] | ||
To see how many molecules from the set are covered by the provided parameters, simply pass '''NEEMP''' those two files (options <code>--sdf-file</code> and <code>--par-file</code>). '''''Figure 4''''' shows a close-up from the <code>cover</code> mode output for '' | To see how many molecules from the set are covered by the provided parameters, simply pass '''NEEMP''' those two files (options <code>--sdf-file</code> and <code>--par-file</code>). '''''Figure 4''''' shows a close-up from the <code>cover</code> mode output for ''set01.sdf'' and the parameters set ''Element.par'', both present in the ''examples'' directory (for details on the syntax and the complete output, see the [[NEEMP:Examples#Example 7 - Coverage validation | examples]] or [[NEEMP:Options | option list]] paragraph) | ||
<br style="clear:both" /> | <br style="clear:both" /> | ||
| Line 85: | Line 85: | ||
'''''<u>BASIC USAGE:</u>''''' ''./neemp -m quality --sdf-file examples/set01.sdf --chg-file examples/set01.chg --par-file examples/Element.par | '''''<u>BASIC USAGE:</u>''''' ''./neemp -m quality --sdf-file examples/set01.sdf --chg-file examples/set01.chg --par-file examples/Element.par | ||
To assess the quality of the EEM parameters on a different set than one they were based on, '''NEEMP''''s <code>quality</code> mode can be | To assess the quality of the EEM parameters on a different set than one they were based on, '''NEEMP''''s <code>quality</code> mode can be called. | ||
Required input files are: ''EEM parameters'' (passed as argument of option <code>--par-file</code>) and a ''new set of molecular structures'' | Required input files are: ''EEM parameters'' (passed as argument of option <code>--par-file</code>) and a ''new set of molecular structures'' for which ''ab-initio charges'' have been computed (arguments of respectively <code>--sdf-file</code> and <code>--chg-file</code>). | ||
To redirect to a file helpful information about the per-atom ''QM vs EEM'' charge comparison, utilize <code>--chg-stats-out-file filename</code> option. As describe in the next section this file is required for the generation of the charge correlation graphs and the quality assessment reports (see [[NEEMP:Reports | here]]). | |||
For details on the syntax and additional output, see the [[NEEMP:Examples#Example 8 - Quality validation | examples]] or [[NEEMP:Options | option list]] section. | |||
Latest revision as of 14:02, 29 June 2016
NEEMP features three modes: calculation, parametrization and validation. Each mode can be selected specifying one of the following keywords after the option --mode or -m:
Calculation mode : charges (calculate EEM charges)
Parametrization mode : params (calculate EEM parameters)
Validation mode : cover (display how many molecules are covered by the given parameters set) and quality (perform EEM parameters quality assay)
Additional functionality: NEEMP enables the user to display information about the provided training set, upon calling the keywords -m info or --mode info. Specifically, number of molecules, the total number of atoms and number of atoms for each atomic type are printed; the latter being defined by default as the chemical element and the maximal bond order. For details on the usage syntax, see the examples or option list paragraph.
Calculation mode
[edit]BASIC USAGE: ./neemp -m charges --sdf-file examples/set01.sdf --par-file examples/ElemBond.par --chg-out-file eem_charges
To compute EEM charges with NEEMP, it is necessary to provide the SDF file with molecules for which the charges will be computed (option --sdf-file) and the PAR file with EEM parameters (option --par-file). EEM charges will be written into the file specified by --chg-out-file option.
The user should keep in mind that, because of the EEM method itself, the EEM charges cannot be computed for atoms or atomic types for which the parameters are missing. This situation is well depicted by Figure 1. On the left side the parameter set contains values for all the atomic types in set01.sdf, on the other hand in the right side image it is evident how the same parameter set is lacking values for several atomic types present in set02.sdf, leading to the discard of many molecules (for details on the syntax, see the examples or option list section).
Parametrization mode
[edit]This mode serves for calculation of EEM parameters. Three main components make up the input for this mode:
- a training set of molecules (argument of
--sdf-fileoption)
- a training set of molecules (argument of
- a set of QM charges for each molecule in the sdf file (argument of
--chg-fileoption)
- a set of QM charges for each molecule in the sdf file (argument of
- the specification of the parametrization approach (by
-por--params-method) and its specific options.
- the specification of the parametrization approach (by
Linear Regression (LR)
[edit]BASIC USAGE: ./neemp -m params -p lr-full --sdf-file examples/set01.sdf --chg-file examples/set01.chg --par-out-file new_parameters.par
LR (--params-method lr-full or -p lr-full) is the default parametrization approach in NEEMP, since its application in the parametrization process is well documented and described in literature. The main extension in respect to previous version is the ability for the user to define by which metrics the best performing parameter set will be selected (see figure):
-sR2or--sort-byR2to select the parameter set with the highest squared Pearson coefficient (R2)
-sRMSDor--sort-byRMSDto select the parameter set with the lowest atomic type root mean square difference (avg(RMSDa))
Additionally, the quality of the parameter set may benefit from the usage of a discarding procedure (-d simple or --discard simple). In fact it has been proved that using a subset of the original training set often leads to EEM charges that better agree with the reference QM charges. For useful examples on the LR parametrization approach and the discarding procedure see respectively examples 3/4 and 5. In particular the latter introduces the --limit-iters and --limit-time options, which modulate the behaviour of the discarding procedure, setting respectively an upper bound in the number of iterations and/or in the execution time.
Differential Evolution + Minimization (DE-MIN)
[edit]BASIC USAGE: ./neemp -m params -p de --sdf-file examples/set01.sdf --chg-file examples/charges.chg --par-out-file new_parameter.par
In parallel we developed an advanced EEM parametrization approach specifically tailored to cope with heterogeneous data, which in the past presented quite a challenge in the EEM parametrization framework. It consists in the combination of differential evolution method with a local minimization technique (NEWUOA), hence the acronym DE-MIN. To call this approach in NEEMP use the option --params-method de or -p de.
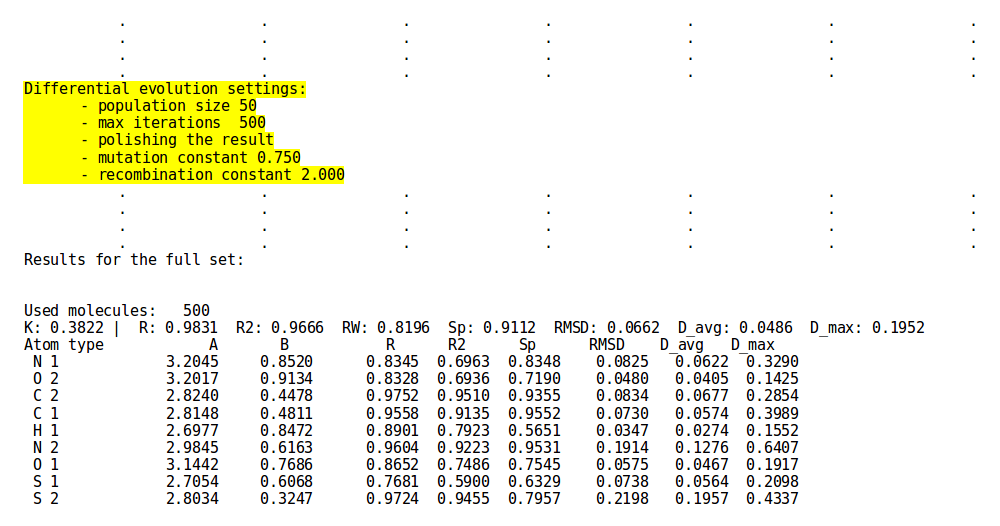
The behaviour of the DE-MIN approach can be modulated by several options, but to make life easier we provide the user with reasonable default values that have been tuned to output optimal parameter set (for a complete view of the options refer to the option list and the examples section).
NB: the default quality evaluating metrics for DE-MIN approach is (avg(RMSDa)).
Validation mode
[edit]This mode allows us to perform two types of EEM parameter set validation - coverage validation and quality validation.
Coverage validation
[edit]BASIC USAGE: ./neemp -m cover -sdf-file examples/set01.sdf --par-file examples/Element.par
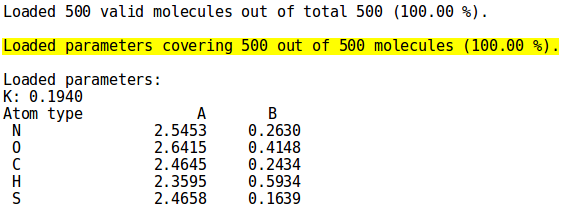
To see how many molecules from the set are covered by the provided parameters, simply pass NEEMP those two files (options --sdf-file and --par-file). Figure 4 shows a close-up from the cover mode output for set01.sdf and the parameters set Element.par, both present in the examples directory (for details on the syntax and the complete output, see the examples or option list paragraph)
Quality validation
[edit]BASIC USAGE: ./neemp -m quality --sdf-file examples/set01.sdf --chg-file examples/set01.chg --par-file examples/Element.par
To assess the quality of the EEM parameters on a different set than one they were based on, NEEMP's quality mode can be called.
Required input files are: EEM parameters (passed as argument of option --par-file) and a new set of molecular structures for which ab-initio charges have been computed (arguments of respectively --sdf-file and --chg-file).
To redirect to a file helpful information about the per-atom QM vs EEM charge comparison, utilize --chg-stats-out-file filename option. As describe in the next section this file is required for the generation of the charge correlation graphs and the quality assessment reports (see here).
For details on the syntax and additional output, see the examples or option list section.