PatternQuery:Explorer: Difference between revisions
Appearance
No edit summary |
|||
| Line 2: | Line 2: | ||
# Querying user submitted data (up to 100 structures). | # Querying user submitted data (up to 100 structures). | ||
# Query smaller subsets of the Protein Data Bank | # Query smaller subsets of the Protein Data Bank | ||
# ''' | # '''Tweaking queries before mining the PDB database'''. | ||
The last point would be probably of the most interest for the wide-range of users. In the following visual you will learn how to work with '''MotiveQuery Explorer'''. | The last point would be probably of the most interest for the wide-range of users. In the following visual you will learn how to work with '''MotiveQuery Explorer'''. | ||
Revision as of 22:08, 28 December 2014
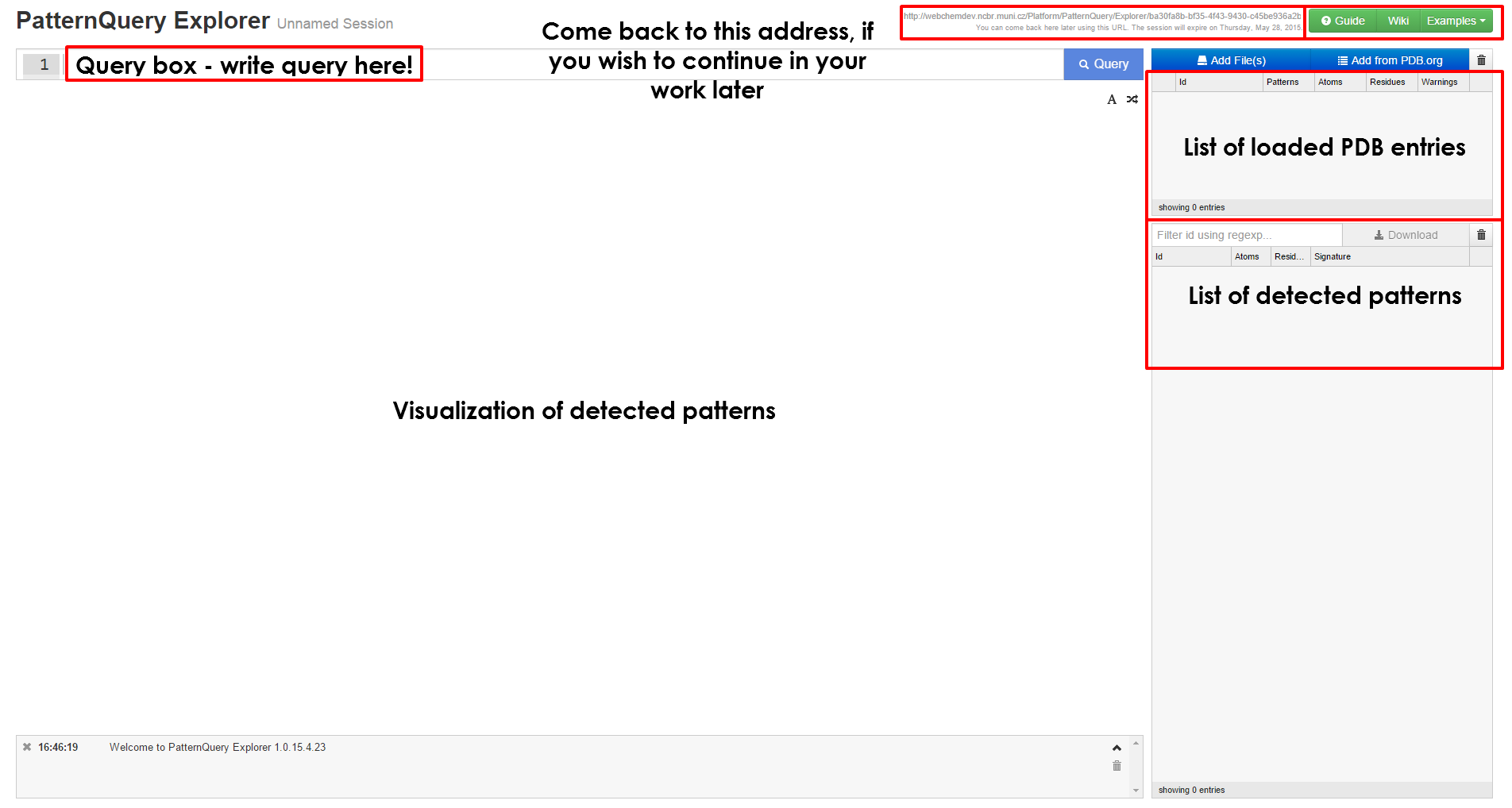
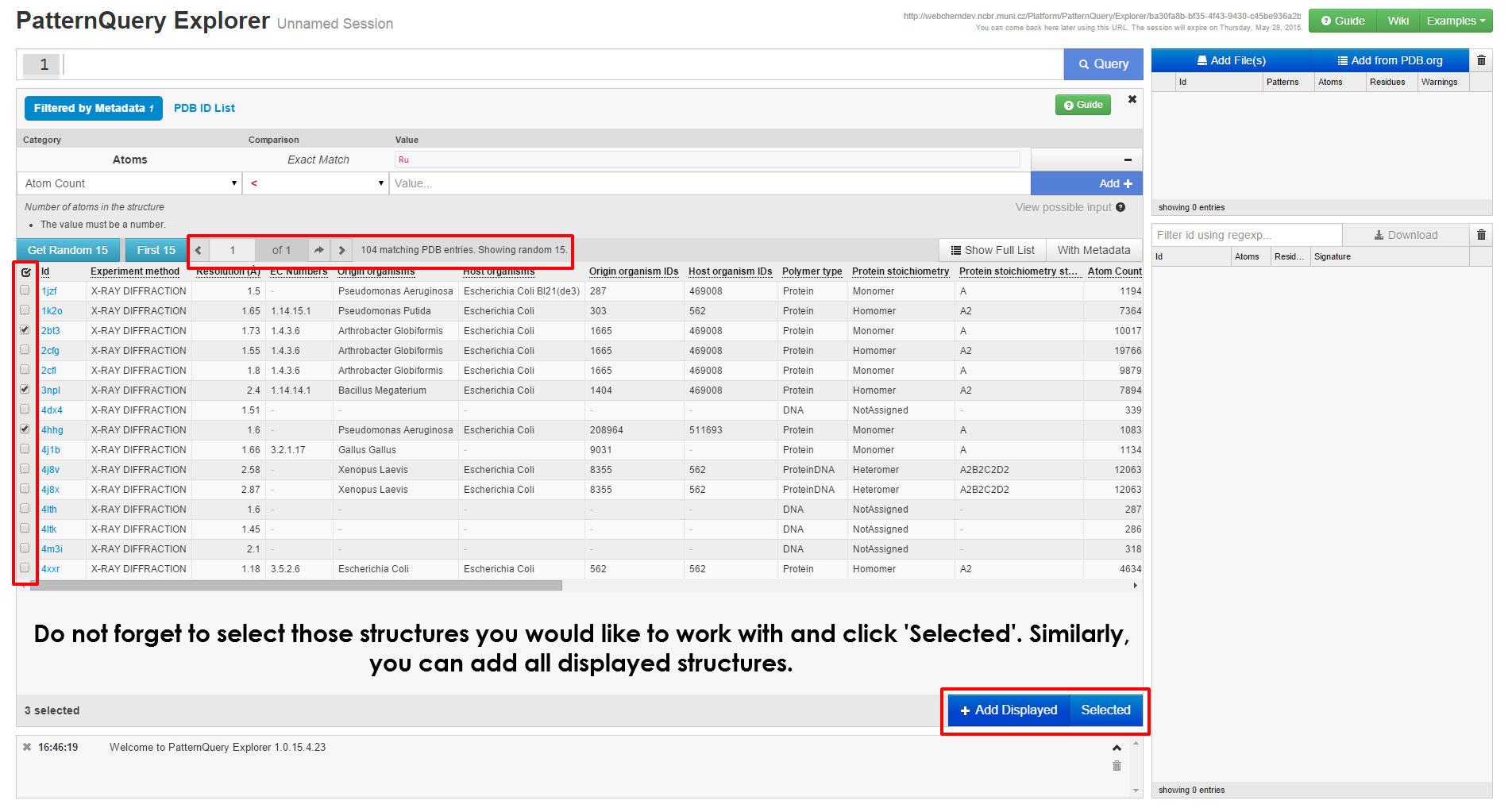
When mining the whole Protein Data Bank for structural fragments it is always better to look before you leap. This is one of the purposes of the MotiveQuery Explorer application. This JavaScript/HTML is tailored for:
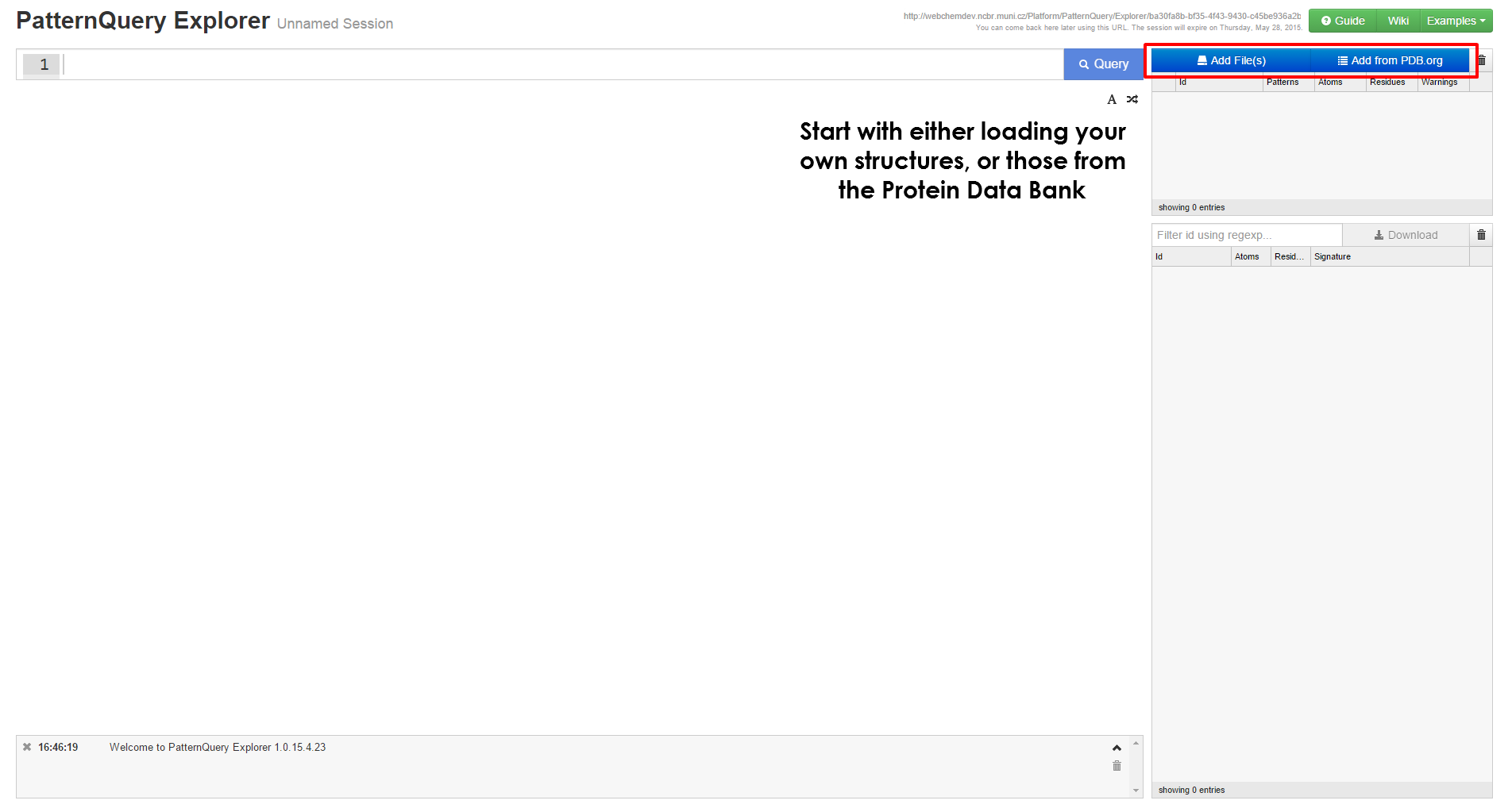
- Querying user submitted data (up to 100 structures).
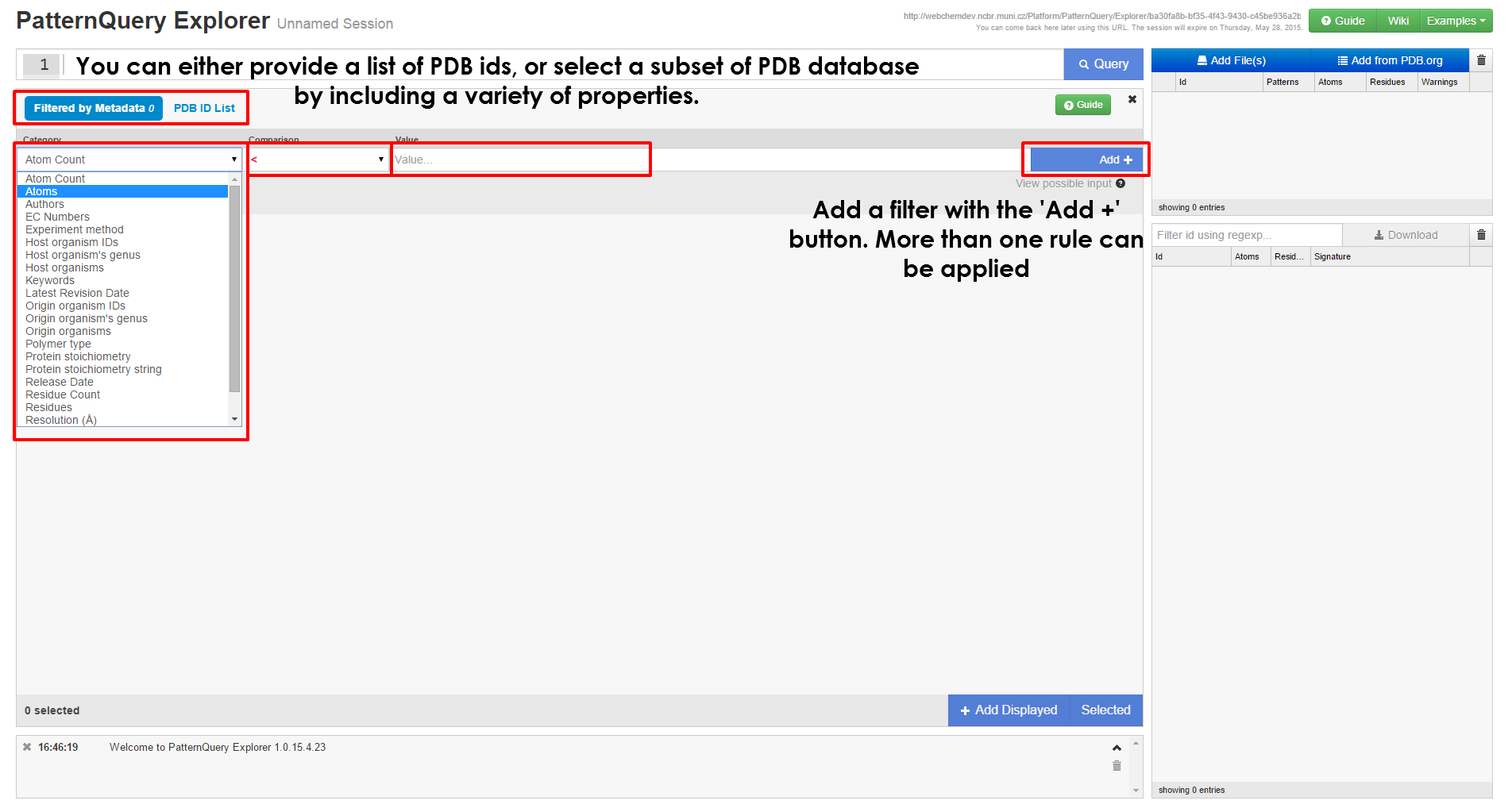
- Query smaller subsets of the Protein Data Bank
- Tweaking queries before mining the PDB database.
The last point would be probably of the most interest for the wide-range of users. In the following visual you will learn how to work with MotiveQuery Explorer.
If the service is not working the way it should, please check your browser compliances in the technical details section.
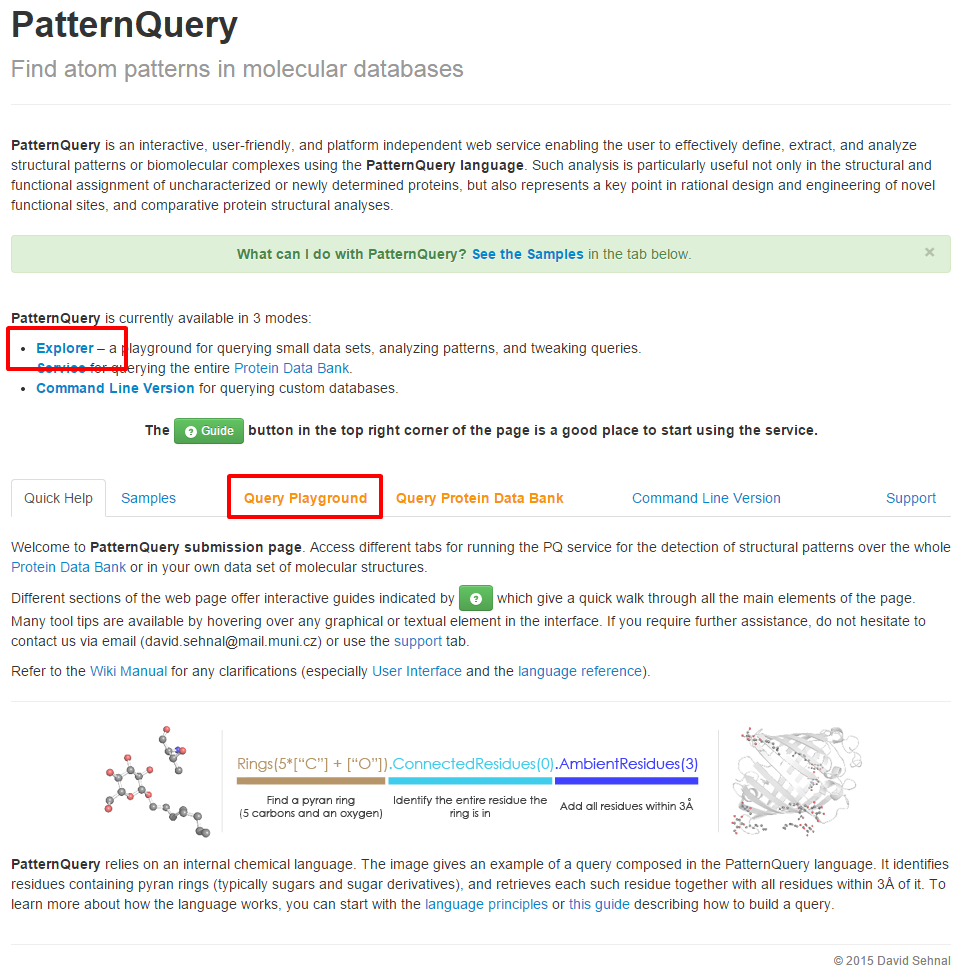
Launch MotiveQuery Explorer app
|
|
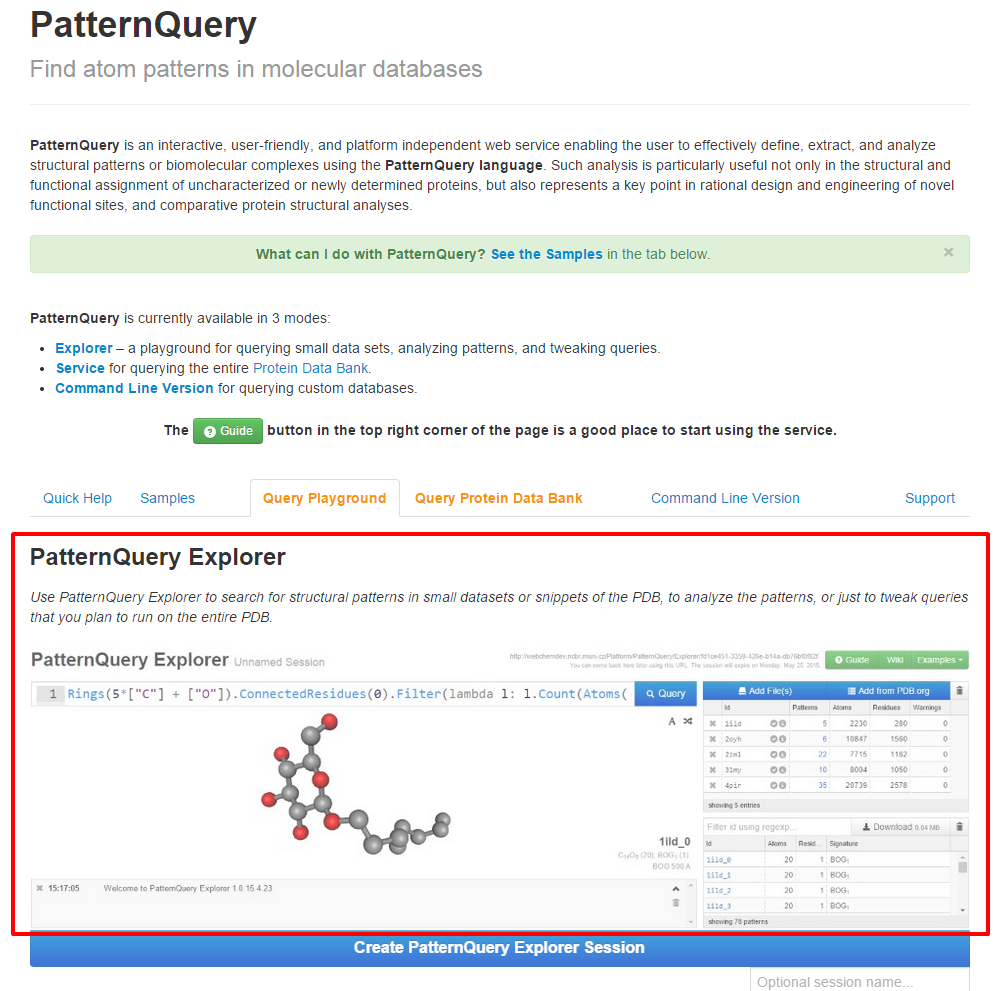
| Follow either Explorer link or Query Playground tab at the Synopsis page, in order to launch the application. | Create a new session or load the previous one. |
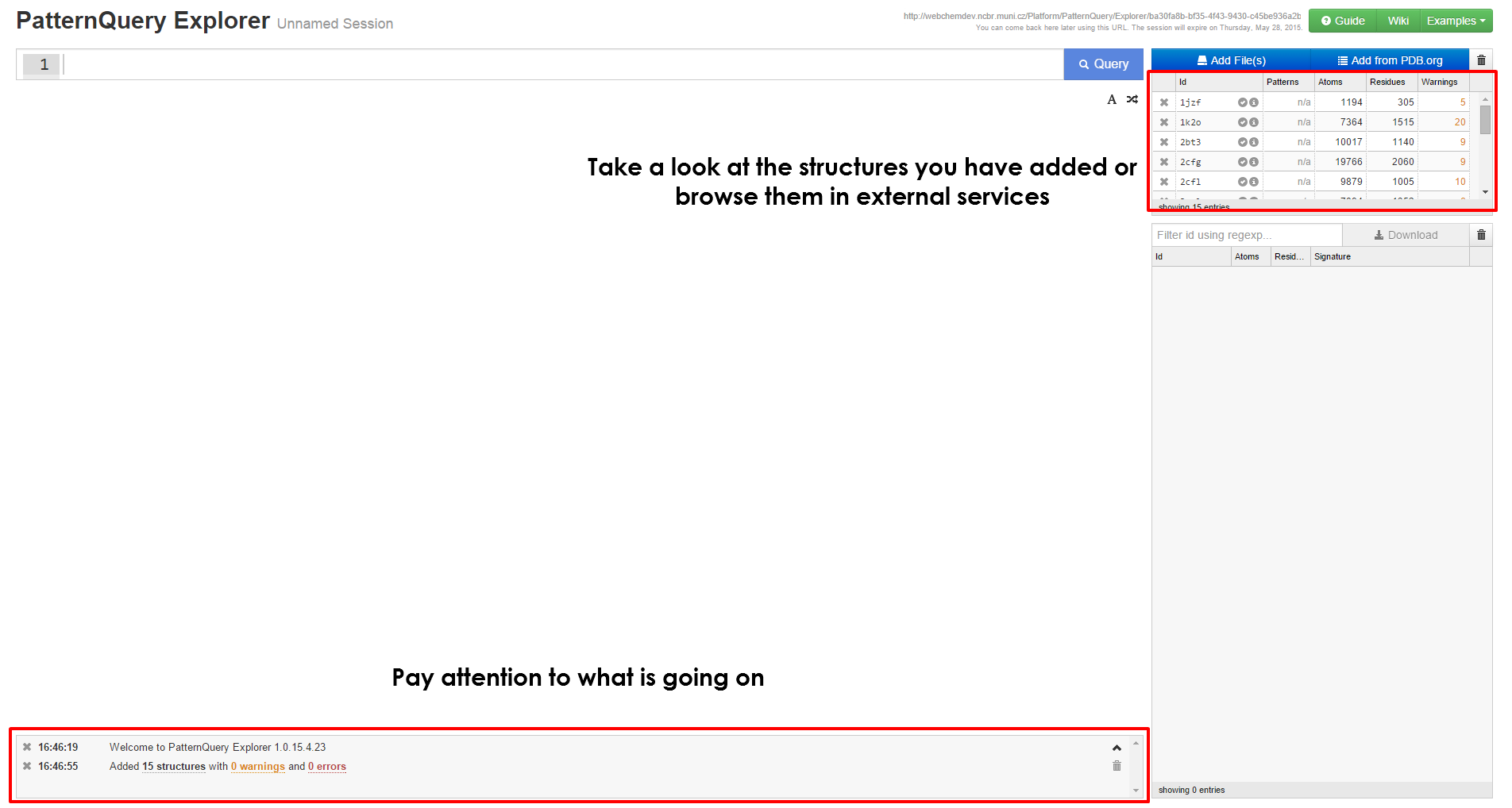
Get started with MotiveQuery Explorer
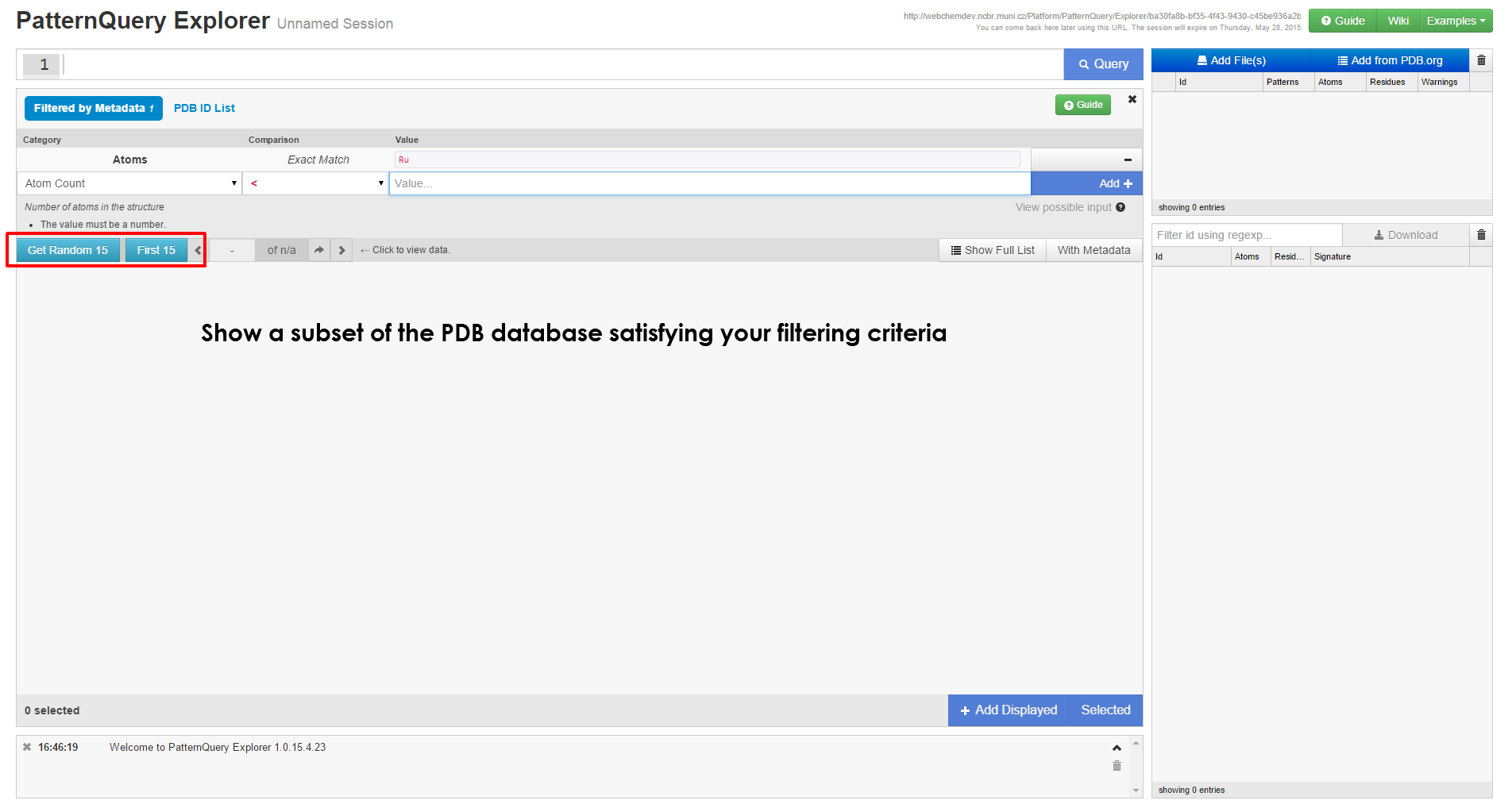
Executing Query & interpreting results
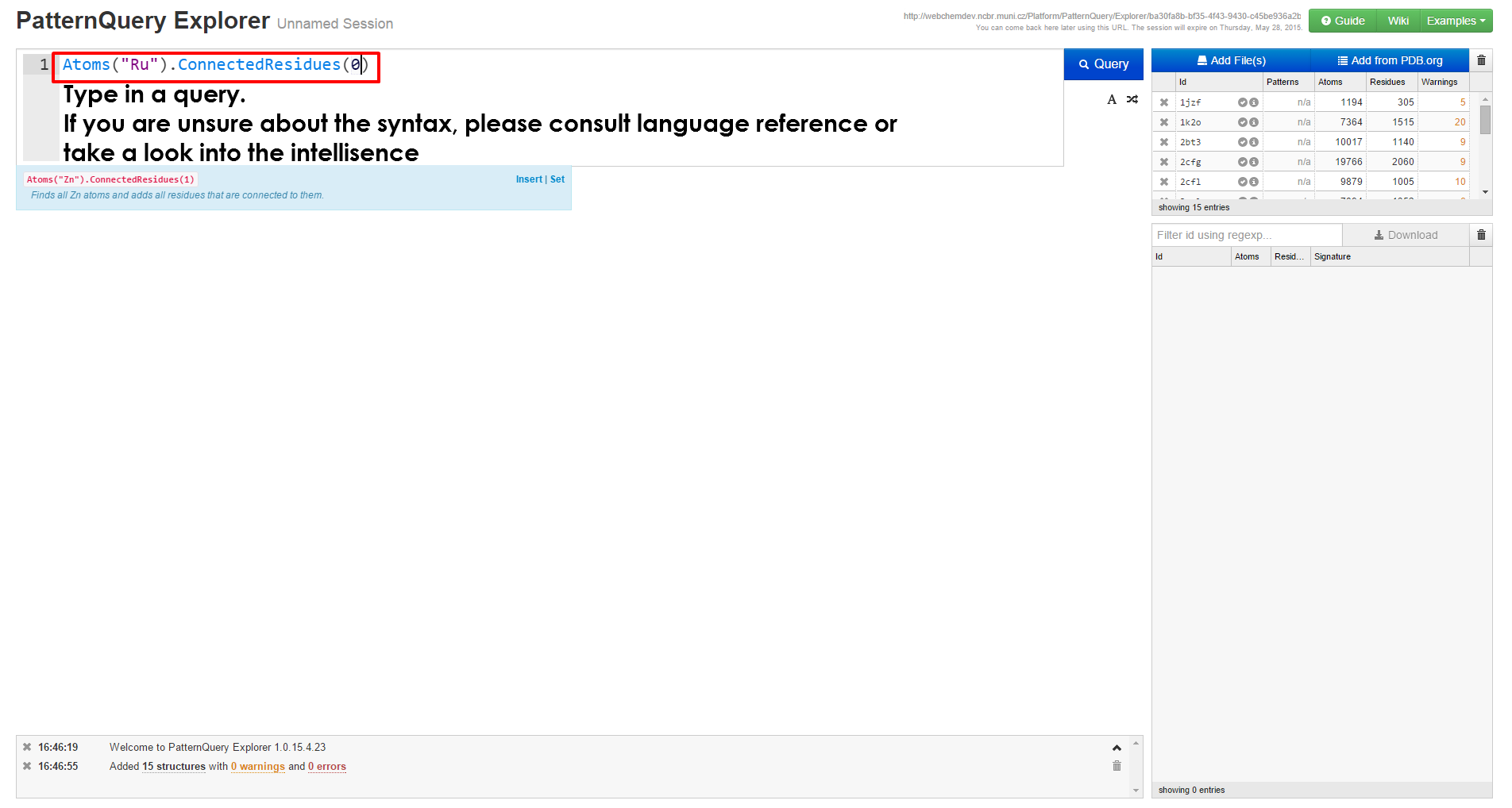
|
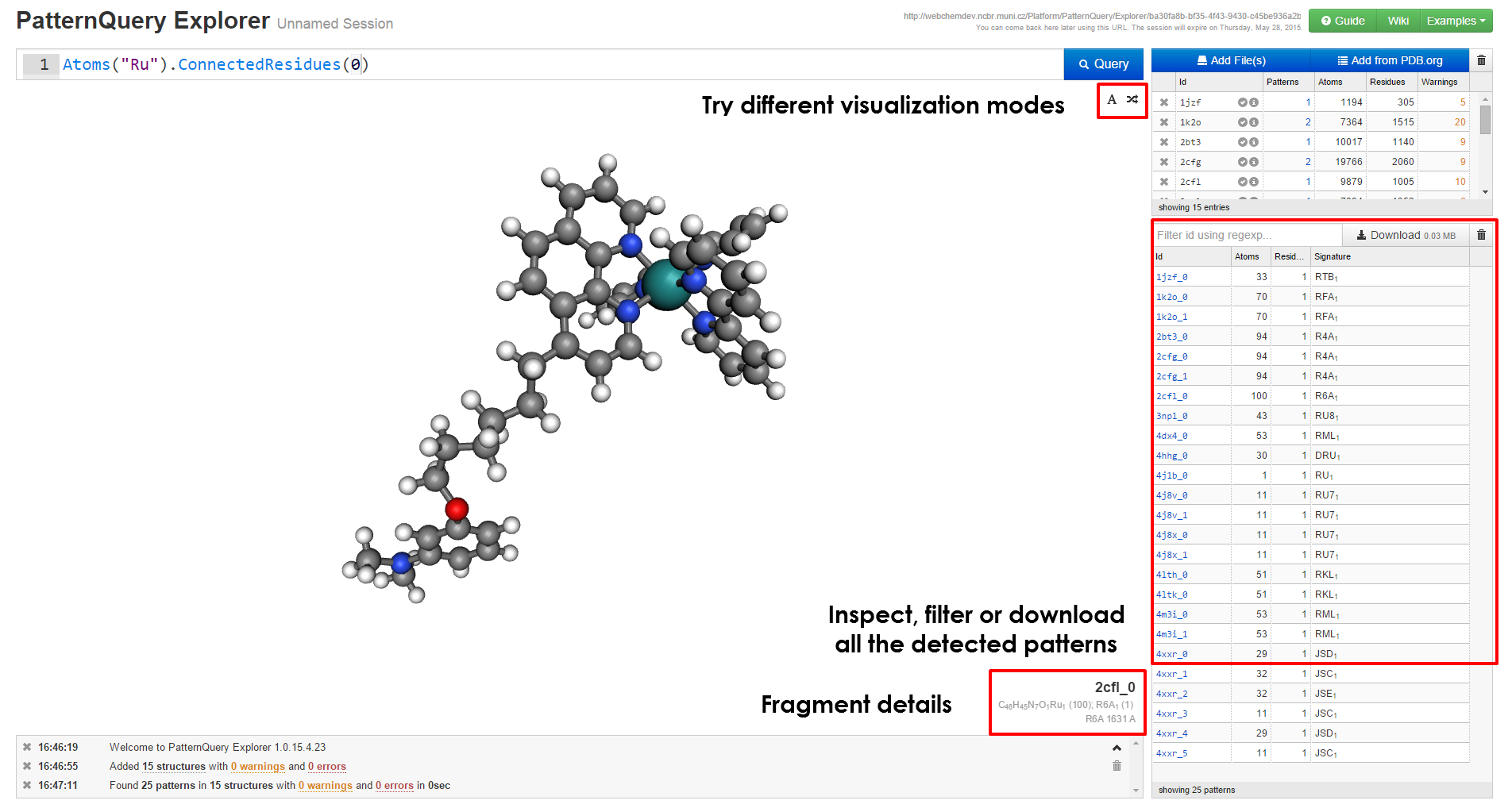
|
| When you have loaded at least a single structure, you can immediately start typing queries in the query box at the top of the page. If you are unsure on how to create query, first browse the principles behind the MotiveQuery language and learn how to build a query. Case studies and language reference might come handy as well. | After the query is executed, the results are visualized in no time, you can inspect them in the browser or download them for further processing. You are free to come back to this web address later, the results are kept for at least a month. |